CmpSpots Reference Manual

CmpSpots Reference Manual
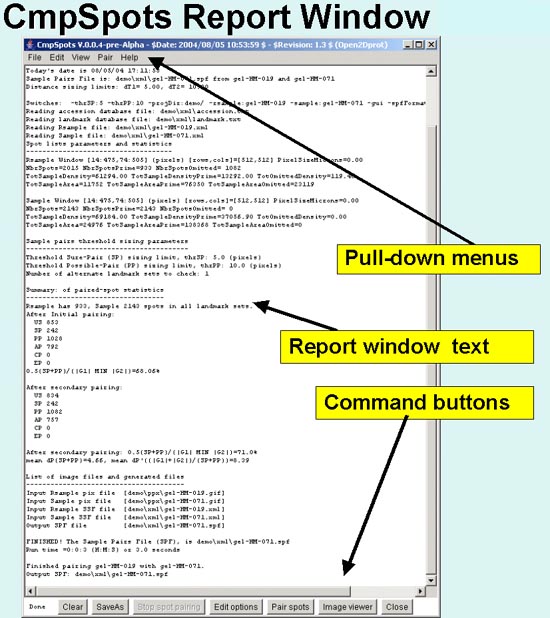
Figure 1. Screen view of the CmpSpots program Report Window interface
This screen shot shows the status of CmpSpots in the
Report Window graphical user interface after a spot pairing of the
demo samples which are 2D PAGE gels. The reference sample or Rsample
is gel-HM-019. The other sample denoted Sample is gel-HM-071) has
completed. The pairing threshold parameters are shown at the top with
the list of Switches. The summary statistics on the spot list
pairing is summarized in the window including the run time [Run on a
1Ghz Pentium-4 Windows 2000 system]. The Report window has five pull-down menus at the top of the window used to
invoke menus: File, Edit, View, Pair, and Help
operations. Although, the button commandss at the bottom of the
window are also available in the menus, they are replicated as buttons
for convenience. The Clear button will clear the window.
SaveAs will let you save the Report Window text in a text file.
The Edit options button pops up a window to let you edit the
options including the sample name, computing window, sizing thresholds
etc. The Pair spots button starts the spot pairing on the
current options. You can stop the spot pairing by pressing the Stop
spot pairing button. After you have paired the spot lists, you can
review the paired spot data by pressing the Image Viewer to
popup the image viewing window. A status area appears in the lower
left corner and reports the current state of the spot pairing during
an analysis. It shows "Done" since the spot pairing had completed.
1. Introduction
CmpSpots is an open source Java 2D spot list matching program for
finding and pairing spots between the two quantified spot lists. It
is part of the Open2Dprot project (
http://open2dprot.sourceforge.net/). CmpSpots is derived the the
GELLAB-II 2D gel spot pairing program cmpgl2 described in
[3]. CmpSpots has been generalized to other types of samples besides
2D gels. While the original program was written in C, CmpSpots is
written in Java, uses XML input and output files and has an optional
graphical user interface. This initial open-source CmpSpots program
code could be used as the basis for more advance spot pairing
methods.
The program may be run either interactively (-gui) or under an OS shell command line interface to implement batch (-nogui). If the default -gui mode is used, after the spot pairing is finished, the user has the option of interactively examining the paired spot data overlayed on the original sample images. The user may also modify the input switch options and save the new options in a "CmpSpots.properties" file in the current project directory when they exit so that the last used options may be used as the default switch options in subsequent running of CmpSpots.
In the analysis of 2D image samples (whether real or virtual), the matching of spots between these samples by pairing spots between their spot lists is difficult to perform manually when there are more than a few spots. Automation is necessary when the samples contain a large number of spots, spots which are at best only locally congruent from sample to sample, when the spots can not be counted on to maintain their "shape" or density and contain little infrastructure on which to build a characterization [3]. There is at best a local congruence between two samples related by some a priori undetermined affine transformation. Some of this description is derived from [3].
One method would be to warp (using a non-linear affine transform) one sample image to the geometry of the reference sample image. Then when these images are analyzed by quantifying segmentmented spots, the spots will be congruent. This method won't work if there are no images (e.g., the 2D spot lists are derived from non-image data (e.g., 2D LC-MS data).
Another method would do a similar warping, but using only the spot lists themselves.
A landmark spot may be defined in various ways. In one current empirical procedure for choosing landmarks, it is a morphologically distinctive spot such that neighboring spots and the landmark spot form a consistent morphological structure. Moreover, this structure should be easily recognized across the set of samples used in an experiment consisting of a number of gels (> 2). The landmark spots are selected to cover the regions of interest of the sample fairly evenly if there is uniform distortion between samples. If there are some regions with a lot of distortion, more landmarks should be used in these regions. Depending of the similarity and distortions between the samples, a few to a larger number may be required. This set of landmarks is called the landmark set.
The probability of finding the same spot in two samples relative to the aligned images of a closeby landmark is greater than if the entire sample spot space were to be searched. This partitioned search has the added advantage that landmark regions contain an order of magnitude fewer spots than the total sample space. Thus the combinatorics of performing the spot matching is greatly decreased as well.
Landmarks could be supplied either using a manual "landmark definition" interactive program (not described here) or might be generated using methods for discovering a subset of robust paired-spots that might be putative landmarks.
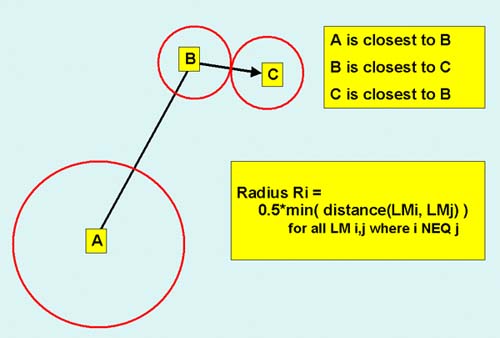
Figure 2. Definition of the effective-radii of a set of landmarks.
The effective-radii of certainty of a landmark Ri is 1/2 the minimum
distance from landmark i to its nearest adjacent landmark j. In this
example, radius Ra > Rb and Rb=Rc. The nearest neighbor of landmark A
is B and its next nearest neighbor landmark is C. Estimates of spot
pairs within landmark radii are more reliable.
Implementation of landmark-oriented spot pairing between two samples
The spot pairing algorithm is described below.
The actual pairing is performed in two passes through the landmark
sets data called the primary and gelssecondary
pairing procedures. Each procedure operates on one landmark set at a
time.
In the primary pairing algorithm (figure 3), the spots are first mapped to the Cartesian coordinate system defined by making the landmark spot (0,0) relative to the origin in the two samples: Rsample (G1) and Sample (G2). Each spot in G1 is provisionaly paired to the spot that is its nearest neighbor in the projected image of G2. Because of possible asymmetry of the two sets the reverse comparison is also performed so that each spot in G2 is provisionally paired with its nearest neighbor spot in Rsample. The nearest neighbor distance is called dP (pair distance). The distance dL is the distance from the landmark spot to the mean locus of the two spots in the provisional pair. Two parameter distances are empirically defined: dTsp and dTpp. Spots closer than dTsp are relatively well paired. Spots greater than dT2 are very poorly paired and possibly should not be paired. The default values of dTsp and dTpp (5 and 10 pixels respectively) were determined empirically, by examination of the nearest neighbor values of several sets of paired samples under a wide variety of conditions. Figure 4 shows various cases which can occur. Four types of pairing labels can be assigned. There are sure pair "SP", possible pair "PP", ambiguous pair "AP" and unresolved spot "US". The primary spot pair labeling assignments are defined in figure 3.
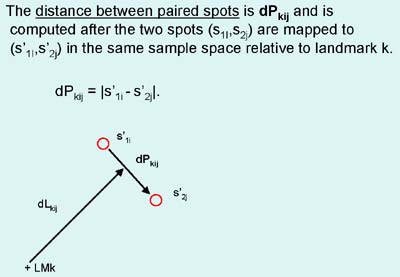
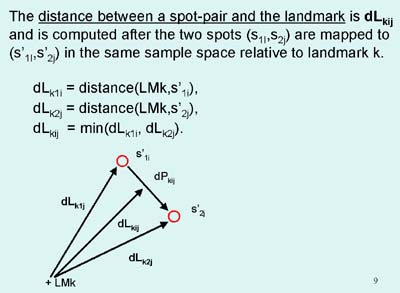
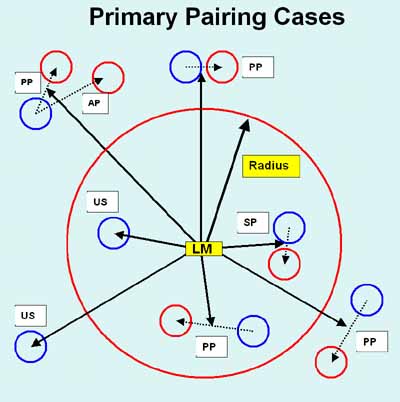
Figure 3. Spot pairing rules.
Spot pairing is peformed using several features of landmarks and spots
in these landmark sets. A) Shows the definition of dP or the
distance between spots of a spot pair mapped to the same space.
B) Shows the definition of dL or the distance from a landmark
to the spot pair mapped to the same space. C) Spot pair
primary labeling assignment definitions. Each potential nearest
neighbor spot pair in a landmark set has one of four labels: SP - sure
pair, PP - possible pair, AP - ambiguous pair, US - unresolved
spot. Rk is the effective radius for a landmark set k. The dTsp is
the threshold distance for SP spots -thrSP: value. The dTpp is
the threshold distance for PP spots -thrPP: value. primary pairing is described below.
The labeling cases are defined by the following cases:Secondary spot pairing optimization
A second pass through the data is peformed to optimize the spot
pairing. Secondary spot pairing can be used to further resolve AP and
US labels in adjacent landmark sets into SP or PP labels which are
then placed in either of the two sets. There are four cases: (a) two
unresolved spots (US and US), (b) two ambiguous pairs (AP and AP),
(c-d) one ambiguous spot (AP) and one unresolved spot (US). The new
spot pair is upgraded to a SP or PP pairing label and is put into
whichever landmark set has the smallest dL for the putative pair. Each
landmark set is tested. Each spot has a list of adjacent landmarks, so
these are tested to see if pairing can be optimized for that spot. The
secondary pairing algorithm
is described below.
The application looks up the sample in the accession database (in
xml/accession.xml or as specified using the -accessionFile switch) and
gets additional information about the sample. The Open2Dprot
http://open2dprot.sourceforge.net/Accession pipeline module is
used for entering samples into the accession database.
[Status: The Open2Dprot Accession module program is not released yet.
The accession database could be edited manually as either XML
(accession.xml), or tab-delimited text (accession.txt) with Excel.]
The samples correspond to Sample Spot-list Files (SSF) and should be
found in the user-project-directory/xml/ sub-directory. The
SSF format is described in the Open2Dprot Seg2Dgel
program. SSF files may be used by CmpSpots as either XML or full
tab-delimited data formats.
There may be images associated with the samples that can be used with
the Image Viewer. This is the case if the spots were derived from real
images (e.g., 2D gels) or virtual images (e.g., from synthesized 2D
LC-MS data), etc. These images may be in TIFF (.tif, .tiff), JPEG
(.jpg), GIF (.gif), or PPX (.ppx GELLAB-II) format. TIFF images may be
8-bits/pixel through 16-bits/pixel, whereas JPEG, GIF, and PPX are
8-bit images. Gray values in the image files have black as 0. This is
mapped to 0 for white and the maximum pixel value for black.
The input sample image files, if any, are kept in the
user-project-directory/ppx/ sub-directory. This database
directory structure is consistent with and is used by the other
Open2Dprot analysis pipeline programs.
The data output file is called the Sample Pairs File (SPF) and is
saved in the user-project-directory/xml/ directory. The
generated name the same as the base name of the image file but with a
different extension depending on the output format. The possible
extensions are: .spf (for ASCII format compatible with GELLAB-II),
.xml (XML format), and .txt (tab-delimited format). One of these
formats are specified specified by the -spfFormat:{F | G
| T | X} command line switch.
[STATUS: -spfFormat:X is the default. Note that the XML
generated will change with changes in MIAPE.]
The computing window is defined as [x1:x2, y1:y2]. You can set the
computing window using the -cw:x1,x2,y1,y2 command line
switch. If you have not defined it or it is not defined in the
accession database (if the -accessionFile option is used), it is
defined as [0:pixWidth-1 x 0:pixHeight-1] where the virtual image is
of size pixWidth x pixHeight.
[STATUS: The computing window for each sample entry is defined in the
accession database. The Accession program will allow users to define
the computing window.]
If you specify an image to be semented, it will check whether it is in
a ppx/ subdirectory. If not, it will ask you if you want to create a
project directory and will then set up the following four directories
and copy your image into the ppx/ directory. You can also use the
-projDir:user-project-directory switch to specify a (possibly
new) project directory.
If you are using the Windows CmpSpots.exe file or clicking on the
CmpSpots.jar file, you can't change the default startup memory.
However, if you are using the CmpSpots.jar in a script using the java
interpreter as in the following example which uses the -Xmx256M
(specifying using 256Mbytes at startup). Change 256 to a larger size
if you want to increase startup memory.
All logged output is sent to the report window in a scrollable text
window that may be saved or used for cut and paste operations. A set
of command buttons at the bottom of the window are replicates of
commands in the menus, but are easier to access. They include the
following functions:
The documentation is kept on the Internet at
http://open2dprot.sourceforge.net/CmpSpots. Normally, these help
commands should pop up a Web browser that directly points to the
CmpSpots Web page. If your browser is not configured correctly, it
may not be able to be launched directly from the CmpSpots
program. Instead, just go to the Web site with your Web browser and
look up the information there.
An Internet connection is required to download the program from the
Open2Dprot CmpSpots Web site. New versions of the program and
associated demo data will become available on this Web site and can be
uploaded to your computer using the various
(File | Update | ...) menu commands. We currently distribute CmpSpots
so that it uses up to 256Mb. See discussion on
increasing memory.
You can these images in the list below or view all of the screen shots in a
single Web page.
Command line scripts to run the GUI version on one sample are in the
second example and
forth example. These may be downloaded
from
demo-CmpSpots.bat and
demo-CmpSpotsGUI.sh.
The data for these scripts is in the demo/ directory available with the
installation. Alternatively, you can download the demo data from the
Files Mirror as Demo.Z.
The installation packages are available from the
Files mirror
under the CmpSpots releases.
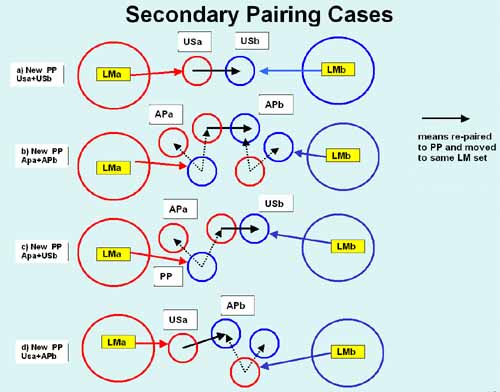
Figure 4. Secondary spot-pairing rules.
A second pass through the data is peformed to optimize the
pairing. Secondary spot pairing can be used to further resolve AP and
US labels in adjacent landmark sets into SP or PP labels which are
then placed in either of the two sets. There are four cases: (a) two
unresolved spots (US and US), (b) two ambiguous pairs (AP and AP),
(c-d) one ambiguous spot (AP) and one unresolved spot (US). The new
spot pair is upgraded to a SP or PP pairing label and is put into
whichever landmark set has the smallest dL for the putative pair. Each
landmark set is tested. Each spot has a list of adjacent landmarks, so
these are tested to see if pairing can be optimized for that spot.
Project directory structure for Open2Dprot and CmpSpots
All Open2Dprot programs assume a project directory structure. This
must exist for the program to proceed. You can either create the
structure prior to running any of the programs or you can create it on
the fly using the -projDir:user-project-directory. It will
lookup and/or create the following sub-directories inside of
theuser-project-directory.
batch/ directory holding temporary batch files - [NOT USED by CmpSpots]
cache/ directory holding temporary CSD cache files - [NOT USED by CmpSpots]
ppx/ directory holding your original gel input files
rdbms/ directory holding CSD database RDBMS files - [NOT USED by CmpSpots]
tmp/ directory holding generated sample image files
xml/ directory that holds accession DB, landmark DB,
SSF spot-list files, and generated SPF paired spot list file
generated by CmpSpots
The use of these directories is discussed in the rest of this
document.Input spot list files
The spot lists to be paired are specified by their sample names (e.g.,
plasma27).
The sample and reference samples to be paired is specified by its
image file name using the -sample and -rsample switches with or
without the file extension (e.g., -sample:plasma27.tif or
-sample:plasma27). The file extension is determined by looking up the
image in the ppx/ project subdirectory at run time.Landmark database
A small (5 to 25 depending on the rubber-sheet distortion between
samples) set of corresponding landmark spots is required for the
Sample and Rsample. These are defined in a landmark database
(xml/landmark.xml) that is uses the same sample names as used in the
accession database. The Open2Dprot
http://open2dprot.sourceforge.net/Landmark pipeline module is used
for defining landmark pairs for (Rsample, Sample) into the landmark
database.Output paired-spot list file - the Sample Pairs File (SPF)
The CmpSpots output is a quantified paired-spot list in various ASCII
formats including XML and tab-delimited as well as the historical
GELLAB-II SPF formats.1.1 Investigating spot pairing using the paired-spot Image Viewer
Spot pairing may be investigated using the Image Viewer button
which pops up the Image Viewer window. This lets you select spots to
review the pairing data including the quantified spot data. You may
add overlays for (subsets of) pairing labels, pairing vectors,
landmarks names, landmarks radii. The current selected spot's landmark
may be used to filter the spots belonging to that landmark set. There
is also a pairing feature histogram that may be used for spot
filtering as well. There are four Image Viewer pull-down menus:
UL>
1.1.1 Image viewer - File menu
These commands are used to change the sample image being displayed.
![]() - specify
the reference sample.
- specify
the reference sample.
![]() Log text to Report Window -
copy all text output from the Image View to the Report window.
Log text to Report Window -
copy all text output from the Image View to the Report window.
------------------------------------
1.1.2 Image viewer - View menu
These commands are used to change the sample image overlays being
displayed.
![]() Add image overlay, else white - add overlay
Add image overlay, else white - add overlay
![]() Add pairing vectors overlay - add overlay
Add pairing vectors overlay - add overlay
![]() Add pairing labels overlay - add overlay
Add pairing labels overlay - add overlay
![]() Add landmarks overlay - add overlay
Add landmarks overlay - add overlay
![]() Add landmark radii overlay - add overlay
Add landmark radii overlay - add overlay
------------------------------------
![]() User mouse drag to select spots - select spots by draging
the mouse over spots rather than clicking on them.
User mouse drag to select spots - select spots by draging
the mouse over spots rather than clicking on them.
------------------------------------
![]() Report all spot pairs for selected spot - report in the text
area all paired spots incling all AP labeled spot pairs for the
selected spot.
Report all spot pairs for selected spot - report in the text
area all paired spots incling all AP labeled spot pairs for the
selected spot.
1.1.3 Image viewer - Filter menu
These commands are used to change the histogram filter.
![]() Filter spots by histogram bin - to enable histogram filtering
Filter spots by histogram bin - to enable histogram filtering
![]() Test spot data GEQ histogram data - otherwise use data less
than the histogram bin data you selected.
Test spot data GEQ histogram data - otherwise use data less
than the histogram bin data you selected.
------------------------------------
![]() -
select histogram filter to use
-
select histogram filter to use
![]() Add spots with Sure-Pair (SP) label - filter by SP labels
Add spots with Sure-Pair (SP) label - filter by SP labels
![]() Add spots with Possible-Pair (PP) label - filter by PP labels
Add spots with Possible-Pair (PP) label - filter by PP labels
![]() Add spots with Unresolved-Spot (US) label - filter by US labels
Add spots with Unresolved-Spot (US) label - filter by US labels
![]() Add spots with Ambiguous-Pair (AP) label - filter by AP labels
Add spots with Ambiguous-Pair (AP) label - filter by AP labels
------------------------------------
![]() Filter spots in current landmark set - else use all spots
in all landmark sets
Filter spots in current landmark set - else use all spots
in all landmark sets
1.1.4 Image viewer - Histogram menu
These commands are used to change the histogram filtering criteria.
![]() -
how to analyze the histogram data
-
how to analyze the histogram data
![]() Dist between spots in a pair - filter by dP or the distance
between the two spots after mapped to the same landmark
Dist between spots in a pair - filter by dP or the distance
between the two spots after mapped to the same landmark
![]() Dist between pair and LM - filter by dL or distance
between the center of the spot pair and their landmark
Dist between pair and LM - filter by dL or distance
between the center of the spot pair and their landmark
![]() Labels SP,PP,US,AP - filter by pairing labels labels
Labels SP,PP,US,AP - filter by pairing labels labels
![]() Landmark set size - filter landmark set size
Landmark set size - filter landmark set size
2. CmpSpots spot pairing algorithm
The spot pairing algorithm is described in two parts. The first part
describes the global processing operations. The second part describes
individual spot pairing operations.Global processing algorithm
2.1 Spot pairing within a landmark set - algorithm
This describes the primary spot pairing within a given landmark
set j. This algorithm is applied for each landmark set in turn. See
Figure 3C which illustrates pairing
geometry for two spots in the same landmark set between the two
samples. The decision rules use the pairing cases listed in that figures legend.
Note the symmetry of the algorithm is such that that each sample views
the other sample the same way.
Note: the Image viewer used with the -gui option lets the user
interactively investigate this data.
2.2 Secondary Spot pairing - algorithm
This describes the secondary spot pairing. Secondary spot pairing is
used to further resolve AP and US labels in adjacent landmark sets
into PP labels that are then placed in either of the two sets. There
are four cases: (a) two unresolved spots (US and US), (b) two
ambiguous pairs (AP and AP), (c-d) one ambiguous spot (AP) and one
unresolved spot (US). The new spot pair is upgraded to a PP pairing
label and is put into whichever landmark set has the smallest dL for
the putative pair. The four cases are described in Figure 4. This procedure is
applied to each landmark set in the Rsample and then each landmark set
in the Sample. Again, the algorithm is symmetric and is applied to
each sample for each landmark set.
3. Running CmpSpots and specifying parameter options via the
command line
The program may be run either interactively (-gui) with a graphical
user interface (GUI) or under an OS shell command to implement batch
(-nogui) depending on how it was started. In the former case, after
the spot pairing is finished, the user has the option of interactively
viewing the paired spot data using the Image Viewer. The user may
also modify the input switch options and save the new options in a
"CmpSpots.properties" file in the current project directory so that it
may be used as the default switch options in subsequent running of the
program.
[Status: the CmpSpots.properties file is not enabled.]
All options including the input reference sample and other sample to
be paired are specified via GNU/Unix style switches on the command
line (-switch{optional ':parameters'} and its negation as
-noswitch). However, if GUI mode is used, you can interactively
specify the switches and their options. It is assigned previously by
software that generated the SSF spot lists.
The computing window region of interest
The computing window is a rectangular region or interest in the SSF
spot list (real or virtual) image where data is considered to be
valid. Spots in this region should be paired. Any spots outside of
this region are ignored.Local Folders and files created and used by CmpSpots
When CmpSpots is first started, it will check for the following folders
and files in the installation directory and create them if they can
not be found.
CmpSpots command-line arguments switch usage
The command line arguements usage is:
CmpSpots -rsample:Reference-sample -sample:sample [< optional switches >]
The complete list of switches is given
later in this manual and as well as some examples of typical sets of switches. The
user defined default switches may be specified as a resource string
'CmpSpots.properties' file saved in the project directory. For
example:
CmpSpots -rsample:gel-HM-19 -sample:gel-HM-071 -thrSP:5 -thrPP:10 -project:demo/ -gui
Options wizard window for setting the command line switches
If you invoke the Edit options button in the Report window (or
from the Edit menu), it will popup an options wizard shown in Figure 5
to let you set or change the switch options and then to save these as
the new default switch options. The default is saved in the
CmpSpots.properties file when you exit program.
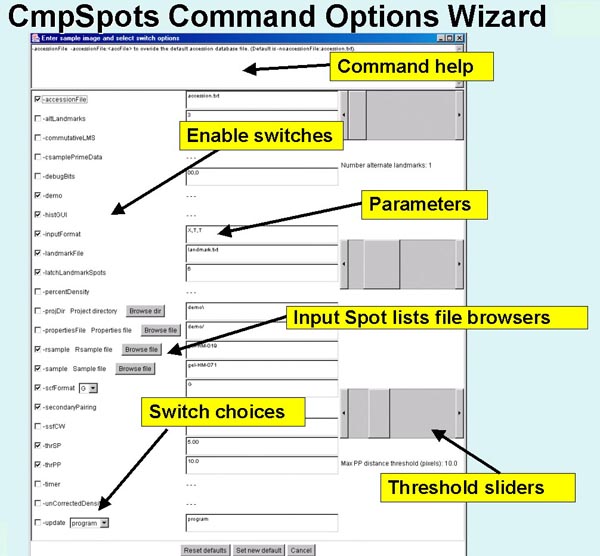
Figure 5. Screen view of the popup options wizard window for
setting the command line switches, parameter and specifying input
samples to be paired.
All of the switches are available in the
scrollable window. Switches are checked if they are enabled and if the
switch requires a value, the current value is shown in the data entry
window to its right. On the right are several threshold sliders for
the the upper sizing values for several parameters including
-thrSP:dTsp, -thrPP:dTpp,
-latchLandmarkSpots:maxLatchDist, and
-altLandmarks:nbrAltLMs. In the middle, are several
Browse buttons to use for specifying a different samples
(-rsample: and -sample:), and directories. Clicking on
any switch will show a short help message associated with that switch
at the top of the window. Pressing the Set new default button
will pass the new options values back to CmpSpots. Note: for this to
take effect, you must exit and then restart CmpSpots. Then to use
them, press the Pair spots button in the main Report Window.
Updating CmpSpots from the Open2Dprot Web server using -update switch
As new versions of CmpSpots are developed and put on the Web server,
a more efficient way of updating your version is to use the -update
commands. There are four options:
-update:program to update the program jar file
-update:demo to update the demonstration files
-update:doc to update the documentation files
-update:all to update all of the above
After updating the program, it should be exited and restarted for the
new program to take effect. Increasing the allowable memory used by CmpSpots
If you are working with very large images that require a lot of memory,
you might want to increase the memory available at startup.
java -Xmx256M -jar CmpSpots.jar {additional command line args}
4. Command and Report Window - the command center
CmpSpots is designed to be used efficiently in a batch mode with
minimal command line output. It is also designed to optionally provide
a graphical user interface (GUI) which creates a Report Window that captures a report
of the spot pairing output as well as additional output directed to it
by the user. There are a set of pull-down
menus as well as a set of buttons for often used functions.
5. Pull-down menus in the Graphical User Interface (GUI)
The menu bar a the top of the Report Window contains five menus.
Menu notation
In the following menus, selections that are sub-menus are
indicated by a '![]() '. Selections prefaced with a '
'. Selections prefaced with a '![]() ' and indicate '
' and indicate '![]() ' indicate that the command is a checkbox
that is enabled and disabled respectively. Selections prefaced with a
'
' indicate that the command is a checkbox
that is enabled and disabled respectively. Selections prefaced with a
'![]() ' and indicate '
' and indicate '![]() ' indicate that the command is a
multiple choice "radio button" that is enabled and disabled
respectively, and that only one member of the group is allowed to be
on at a time. The default values set for an initial database are shown
in the menus. Selections that are not currently available will be
grayed out in the menus of the running program. The command short-cut
notation C-key means to hold the Control key and then
press the specified key.
' indicate that the command is a
multiple choice "radio button" that is enabled and disabled
respectively, and that only one member of the group is allowed to be
on at a time. The default values set for an initial database are shown
in the menus. Selections that are not currently available will be
grayed out in the menus of the running program. The command short-cut
notation C-key means to hold the Control key and then
press the specified key.
5.1 File menu
These commands are used to open the samples to be paired and other
file operations. The current menus and the menu commands (non-working
commands have a '*' prefix) are listed below. You can use either the
"Edit options" button to popup the Options Window editor to change the
input samples or the (File menu | Open Rsample) and (File
menu | Open Sample) commands.
------------------------------------
------------------------------------
![]() - update CmpSpots
programs and data from open2dprot.sourceforge.net/CmpSpots
server
- update CmpSpots
programs and data from open2dprot.sourceforge.net/CmpSpots
server
------------------------------------
5.2 Edit menu
These commands are used to change various defaults. These are saved
when you save the state and when you exit the program.
![]() - change the command
line options
- change the command
line options
5.3 View menu
This menu contains commands to invoke the image viewer used to inspect
the images after spot pairing.
![]() Add histogram to Image
viewer - to add a histogram of spot pairing data to the
Image viewer (do this before starting the Image viewer).
Add histogram to Image
viewer - to add a histogram of spot pairing data to the
Image viewer (do this before starting the Image viewer).
------------------------------------
5.4 Pair menu
This menu is used to run the spot pairing to perform the
analysis. These commands also appear as command buttons at the bottom
of the Report Window.
5.5 Help menu
These commands are used to invoke popup Web browser documentation on
CmpSpots. Some of the commands will load local documentation in the
the GUI report window.
![]() - this
reference manual
- this
reference manual
--------------------------------------
--------------------------------------
6. Downloading, installing and running CmpSpots
The installation packages are available for
download from the SourceForge Files mirror. Look for the most
recent release named "CmpSpots-dist-V.XX.XX.zip". These releases include
the program (both as Windows .exe file and a .jar file), required jar
libraries, demo data, Windows batch and Unix
shell scripts. Download the zip file and put the contents where you want
to install the program. Note that there is a CmpSpots.exe
(for Windows program). You might make a short-cut to this to use in more easily
starting the program. Alternatively, you can use the sample .bat and .sh scripts
to run the program explicitly via the java interpreter. Note that this method
assumes that you have Java installed on your computer and that it is at
least JDK (Java Development Kit) or JRE (Java Runtime Environment)
version 1.5.0. If you don't have this, you can download the latest version free
from the java.sun.com Website.
6.3 Running CmpSpots
There are several ways to run the program. On Windows, you can start
CmpSpots by clicking on the startup icon shown in Figure 6 below.
For Unix systems including MacOS-X, you can start CmpSpots from
the command line by running the CmpSpots.jar file. If your computer is setup
to execute jar files, just type the jar file. In both systems, you can specify
additional command line arguments in Windows .bat and unix .sh scripts (see
demo examples below.
![]()
Figure 6. Startup icon for CmpSpots.exe in Windows.
Clicking on the icon starts CmpSpots. To start CmpSpots, click on the startup
icon shown in Figure 6 below - or you can run the demo-CmpSpots.bat
script. For Unix systems including MacOS-X, you can start CmpSpots from the
command line by clicking on the CmpSpots.jar file or using the
demo-CmpSpots.sh script. These two scripts run the program in batch.
There are also GUI versions of the two scripts demo-CmpSpots-GUI.bat
and demo-CmpSpots-GUI.bat that will pop up a user interface.
You could make short-cuts (Windows) or symbolic-links in Unix to make it
easier to start.
6.4 Requirements: minimum hardware and software requirements
A Windows PC, MacIntosh with MacOS-X, a Linux computer or a Sun
Solaris computer having a display resolution of at least
1024x768. We find that a 1024x768 is adequate, but a 1280x1024 screen
size much better since you can see the Popup Report window, Options
window, and Image Viewer window at the same time. At least 30 Mb of
memory available for the application is required and more is desirable
for comparing large images or performing transforms. If there is not
enough memory, it will be unable to load the images, the transforms
may crash the program or other problems may occur.6.5 Files included in the download
The following files are packaged in the distribution you install.
you can periodically a (File | Update from Web server | ...
program) menu command to update the files from the open2dprot.sourceforge.net
Web server.
You can do a (File | Update from Web server | CmpSpots demo files)
menu command to update it.
7. List of the command line switches
The command line usage is:
CmpSpots -rsample:reference-sample -sample:sample [< optional switches >]
where the order of arguments is not relevant. In the following list,
items in bold are specific values which must be used (e.g., for
-spfFormat:{X | F | T | G}, whereas
variable values in italics indicate that a numeric value for
that variable should be used (e.g., for -thrSP:t1threshold it
might be -thrSP:5). Some switches have several alternate fixed choices
in which case this indicated as a list of bolded items inside of a set
of '{...}' with '|' separating the items. You must pick one of the
items and do not include the '{}' brackets. Also, do NOT include any
extra spaces in the arguments of the switch - it will be counted as if
it were another switch.
Command line switches
-accessionFile:accFile to overide the default accession
database file. (Default is -noaccessionFile:accession.xml).
-altLandmarks:NbrOfLandmarkSets specifies the number of alternate
landmarks to check. (Default is -altLandmarks:3).
-commutativeLMS lets you search the landmark database for sample pairs
where the Rsample and Sample are swapped. (Default is
-commutativeLMS).
-csamplePrimeData indicates that the SSF data are canonical Csample'
data created from replicate samples in the Composite Sample
Database (CSD). (Default is -nocsamplePrimeData).
-debugBits:bits,optLandmarkNbr dumps various conditional
debugging parameters onto the report window as well as the
output SPF file. The debugging is active for all landmarks
unless optLandmarkNbr is specified. The 'bits' are the debug
bits specified as either octal or decimal and enable particular
debugging output if the program was compiled with debugging
enabled. (Default is -nodebug).
-default sets the default switches to a specific configuration:
-nodemo
-thrSP:5 -thrPP:10 -inputFormat:X,X,X
This disables -demo if it was set. (Default -nodefault).
-demo sets the default switches and sample input sample to a specific
configuration. This may be overriden by turning off the -demo
switch in the Options Wizard.
-dtd adds the XML DTD file (Open2Dprot-SPF.dtd) in the output XML
if -spfFormat:X is set. (Default is -nodtd).
-gui to start the spot pairing with a popup Graphical User Interface
rather than in batch mode. This captures messages from CmpSpots.
You can then cut and paste the results or save it to a text
file. The GUI is also used to change the switch options, re-run
the spot pairing and view images after each analysis. (Default
is -nogui).
-histGUI will bring the the dynamic histogram graphical user interface
when the Image Viewer is requested. (Default is -histGUI).
-inputFormat:ssfMode,accMode,lmsMode defines the input formats
for the Sample Spot-list File (ssfMode), accession database file
(accMode), and landmark database file (lmsMode). Where mode is:
F for full tab-delimited data that includes the 3 files:
list of spots, parameters, and statistics. The T for
tab-delimited (.txt) data, and X for XML (.xml)
data. (Default is -noinputFormat:X,T,T).
-landmarkFile:lmsFile to overide the default landmark database
file. (Default is -nolandmarkFile:landmark.xml).
-latchLandmarkSpots:maxLatchDist is the maximum distance to use
when trying to latch a landmark to its closest spot. (Default
is -nolatchLandmarkSpots:6).
-percentDensity to output percent density instead of OD values.
(Default is -nopercentDensity).
-projDir:alternate project directory path to specify the
project directory to use instead of the default 'demo/' file in
the installation directory. (Default is -noprojDir).
-propertiesFile:alternate 'CmpSpots.properties' file to specify
the alternate startup properties file to use instead of the
default 'CmpSpots.properties' file. (Default is
-nopropertiesFile).
-rsample:
7.1 Examples of some typical sets of switches
The following shows a few examples of useful combinations of command
line switches.
Any case-independent switch may be negated by preceeding it with
a 'no' eg. '-notimer'.
The command line syntax used to invoke it is:
CmpSpots input-sample-image-file [< opt.-switches >]
The following examples using switches might be useful:
CmpSpots -rsample:gel-HM-019 -sample:gel-HM-071 -nogui -project:demo/
# Pair two samples into a SPF file using "-spfFormat:X" XML format.
# This is what is normally used in batch mode
CmpSpots -rsample:gel-HM-019 -sample:gel-HM-071 -gui -project:demo/
# Pair two samples into a SPF file, popup up Report window and then
# use may pop up Image viewer to display sample images with paired spot
# overlays including spot labels, vectors and landmark sets
CmpSpots -rsample:gel-HM-019 -sample:gel-HM-071 -spfFormat:G
-project:demo/
# Pair two samples into a SPF file using the GELLAB format.
CmpSpots -rsample:gel-HM-019 -sample:gel-HM-071 -thrSP:10 -thrPP:20 \
-altNbrLMs:2 -project:demo/
# Pair two samples as above, but use new thresholds thrSP, thrPP and
the alternate number of landmarks.
7.2 Debug option bits for the -debug switch
The following are the orthogonal octal -debug option code bits. This
means you can add them together (in octal) and use that computed octal
number (it will also accept decimal). The
-debug:bits,landmarkNbr option is meant only for serious
programmers reading or modifying the source code.
Debugging option bits used with the "-dbug:
8. Demonstrations
8.1 Examples - samples of screen shots
To give the flavor of running the spot pairing program, we provide a
few screen shots of the graphical user interfaces and some images
generated by the program.
8.2 Example - output of the Report Window for a spot pairing
The following Report Window output was generate for the images in the
above example.
CmpSpots V.0.0.4-pre-Alpha - $Date$ - $Revision$ (Open2Dprot)
Today's date is 08/04/04 14:37:34
Switches: -thrSP:5 -thrPP:10 -projDir:demo/ -rsample:gel-HM-019
-sample:gel-HM-071 -gui -spfFormat:X,T,T -spfFormat:G
CmpSpots V.0.0.4-pre-Alpha - $Date$ - $Revision$ (Open2Dprot)
Today's date is 08/04/04 14:37:34
Sample Pairs File is: demo\xml\gel-HM-071.spf from gel-HM-019 and gel-HM-071
Distance sizing limits: dT1= 5.00, dT2= 10.00
Switches: -thrSP:5 -thrPP:10 -projDir:demo/ -rsample:gel-HM-019 -sample:gel-HM-071 -gui -spfFormat:X,T,T -spfFormat:G
Reading accession database file: demo\xml\accession.txt
Reading landmark database file: demo\xml\landmark.txt
Reading Rsample file: demo\xml\gel-HM-019.xml
Reading Sample file: demo\xml\gel-HM-071.xml
Spot lists parameters and statistics
------------------------------------
Rsample Window [14:475,74:505] (pixels) [rows,cols]=[512,512] PixelSizeMicrons=0.00
NbrSpots=2015 NbrSpotsPrime=933 NbrSpotsOmitted= 1082
TotSampleDensity=61294.00 TotSampleDensityPrime=13292.00 TotOmittedDensity=119.40
TotSampleArea=11752 TotSampleAreaPrime=76350 TotSampleAreaOmitted=23119
Sample Window [14:475,74:505] (pixels) [rows,cols]=[512,512] PixelSizeMicrons=0.00
NbrSpots=2143 NbrSpotsPrime=2143 NbrSpotsOmitted= 0
TotSampleDensity=69184.00 TotSampleDensityPrime=37056.90 TotOmittedDensity=0.00
TotSampleArea=24976 TotSampleAreaPrime=138368 TotSampleAreaOmitted=0
Sample pairs threshold sizing parameters
----------------------------------------
Threshold Sure-Pair (SP) sizing limit, thrSP: 5.0 (pixels)
Threshold Possible-Pair (PP) sizing limit, thrPP: 10.0 (pixels)
Number of alternate landmark sets to check: 1
Summary: of paired-spot statistics
----------------------------------
Rsample has 933, Sample 2143 spots in all landmark sets.
After Initial pairing:
US 853
SP 242
PP 1028
AP 792
CP 0
EP 0
0.5(SP+PP)/(|G1| MIN |G2|)=68.06%
After secondary pairing:
US 834
SP 242
PP 1082
AP 757
CP 0
EP 0
After secondary pairing: 0.5(SP+PP)/(|G1| MIN |G2|)=71.0%
mean dP(SP+PP)=4.66, mean dP'((|G1|+|G2|)/(SP+PP))=8.39
List of image files and generated files
---------------------------------------
Input Rsample pix file [demo\ppx\gel-HM-019.gif]
Input Sample pix file [demo\ppx\gel-HM-071.gif]
Input Rsample SSF file [demo\xml\gel-HM-019.xml]
Input Sample SSF file [demo\xml\gel-HM-071.xml]
Output SPF file [demo\xml\gel-HM-071.spf]
FINISHED! The Sample Pairs File (SPF), is demo\xml\gel-HM-071.spf
Run time =0:0:3 (H:M:S) or 3.1 seconds
Finished pairing gel-HM-019 with gel-HM-071.
Output SPF: demo\xml\gel-HM-071.spf
8.3 Examples - part of Sample Pairs File XML format
This is part of a Sample Pairs File to illustrate the type of data
available as output. This used the default -spfFormat:X option
with the XML DTD file Open2Dprot-SPF.dtd.
Open2Dprot-SPF.dtd (if -dtd switch used), else
<?xml version="1.0" ?>
<SpotList_Pairing>
<Pairing_parameters>
<date>"05/10/05 11:18:32"</date>
<Open2Dprot_SPF_Version>"1.5"</Open2Dprot_SPF_Version>
<Project_directory>"demo\"</Project_directory>
<Sample_Pairs_File>"gel-HM-071-SPF.xml"</Sample_Pairs_File>
<thrSP_threshold>5.0</thrSP_threshold>
<thrPP_threshold>10.0</thrPP_threshold>
<nbrAltLandmarks>1</nbrAltLandmarks>
<nbrLandmarks>22</nbrLandmarks>
<Rsample>
<Sample_Type>"Rsample"</Sample_Type>
<Sample_Name>"gel-HM-019"</Sample_Name>
<Simple_FileName>"gel-HM-019.gif"</Simple_FileName>
<Sample_Pix_FileName>"demo\ppx\gel-HM-019.gif"</Sample_Pix_FileName>
<Sample_SSF_FileName>"demo\xml\gel-HM-019-SSF.xml"</Sample_SSF_FileName>
<cwx1>14</cwx1>
<cwx2>475</cwx2>
<cwy1>74</cwy1>
<cwy2>509</cwy2>
<Pix_Height>512</Pix_Height>
<Pix_Width>512</Pix_Width>
<PixelSizeMicrons>0.00</PixelSizeMicrons>
<NbrSpots>1588</NbrSpots>
<NbrSpotsPrime>738</NbrSpotsPrime>
<NbrSpotsOmitted>850</NbrSpotsOmitted>
<TotSampleDensity>8838.70</TotSampleDensity>
<TotSampleDensityPrime>5670.40</TotSampleDensityPrime>
<TotOmittedDensity>91.90</TotOmittedDensity>
<TotSampleArea>36158</TotSampleArea>
<TotSampleAreaPrime>23019</TotSampleAreaPrime>
<TotSampleAreaOmitted>13139</TotSampleAreaOmitted>
<PctOmittedToDprimeAcceptedSpots>1.62 </PctOmittedToDprimeAcceptedSpots>
<Nbr_Spots_Failing_Area_Sizing>
<nbr_below_t1Area_thr>3316 </nbr_below_t1Area_thr>
<nbr_above_t2Area_thr>0 </nbr_above_t2Area_thr>
<nbr_below_t1Density_thr>4214 </nbr_below_t1Density_thr>
<nbr_above_t2Density_thr>0 </nbr_above_t2Density_thr>
<nbr_below_t1Range_thr>0 </nbr_below_t1Range_thr>
<nbr_above_t2Range_thr>0 </nbr_above_t2Range_thr>
</Nbr_Spots_Failing_Area_Sizing>
</Rsample>
<Sample>
<Sample_Type>"Sample"</Sample_Type>
<Sample_Name>"gel-HM-071"</Sample_Name>
<Simple_FileName>"gel-HM-071.gif"</Simple_FileName>
<Sample_Pix_FileName>"demo\ppx\gel-HM-071.gif"</Sample_Pix_FileName>
<Sample_SSF_FileName>"demo\xml\gel-HM-071-SSF.xml"</Sample_SSF_FileName>
<cwx1>6</cwx1>
<cwx2>450</cwx2>
<cwy1>68</cwy1>
<cwy2>503</cwy2>
<Pix_Height>512</Pix_Height>
<Pix_Width>512</Pix_Width>
<PixelSizeMicrons>0.00</PixelSizeMicrons>
<NbrSpots>3059</NbrSpots>
<NbrSpotsPrime>1590</NbrSpotsPrime>
<NbrSpotsOmitted>1469</NbrSpotsOmitted>
<TotSampleDensity>31534.10</TotSampleDensity>
<TotSampleDensityPrime>23052.90</TotSampleDensityPrime>
<TotOmittedDensity>131.30</TotOmittedDensity>
<TotSampleArea>80117</TotSampleArea>
<TotSampleAreaPrime>51342</TotSampleAreaPrime>
<TotSampleAreaOmitted>25905</TotSampleAreaOmitted>
<PctOmittedToDprimeAcceptedSpots>0.5694 </PctOmittedToDprimeAcceptedSpots>
<Nbr_Spots_Failing_Area_Sizing>
<nbr_below_t1Area_thr>2567 </nbr_below_t1Area_thr>
<nbr_above_t2Area_thr>0 </nbr_above_t2Area_thr>
<nbr_below_t1Density_thr>4107 </nbr_below_t1Density_thr>
<nbr_above_t2Density_thr>0 </nbr_above_t2Density_thr>
<nbr_below_t1Range_thr>0 </nbr_below_t1Range_thr>
<nbr_above_t2Range_thr>0 </nbr_above_t2Range_thr>
</Nbr_Spots_Failing_Area_Sizing>
</Sample>
</Pairing_parameters>
<Paired_Spot>
<LandmarkSet>A</LandmarkSet>
<R_spotNbr>188</R_spotNbr>
<R_dxLM>-2</R_dxLM>
<R_dyLM>-25</R_dyLM>
<R_xC>207.0</R_xC>
<R_yC>166.0</R_yC>
<R_merX1>205</R_merX1>
<R_merX2>210</R_merX2>
<R_merY1>163</R_merY1>
<R_merY2>168</R_merY2>
<S_spotNbr>759</S_spotNbr>
<S_dxLM>-2</S_dxLM>
<S_dyLM>-25</S_dyLM>
<S_xC>226.0</S_xC>
<S_yC>166.0</S_yC>
<S_merX1>224</S_merX1>
<S_merX2>229</S_merX2>
<S_merY1>152</S_merY1>
<S_merY2>158</S_merY2>
<PairingCode>P</PairingCode>
<DP>3.0</DP>
<DL>25.1</DL>
<R_area>18</R_area>
<S_area>25</S_area>
<R_dens>1.560</R_dens>
<S_dens>8.352</S_dens>
<R_dPrime>0.826</R_dPrime>
<S_dPrime>3.515</S_dPrime>
<R_volume>1.347</R_volume>
<S_volume>6.484</S_volume>
<R_MaxDens>0.132</R_MaxDens>
<S_MaxDens>0.480</S_MaxDens>
<R_MinDens>0.043</R_MinDens>
<S_MinDens>0.225</S_MinDens>
<R_MeanBkgDens>0.041</R_MeanBkgDens>
<S_MeanBkgDens>0.193</S_MeanBkgDens>
<R_stdDev_X>1.229</R_stdDev_X>
<R_stdDev_Y>1.173</R_stdDev_Y>
<S_stdDev_X>1.292</S_stdDev_X>
<S_stdDev_Y>1.473</S_stdDev_Y>
</Paired_Spot>
<Paired_Spot>
<LandmarkSet>A</LandmarkSet>
<R_spotNbr>190</R_spotNbr>
<R_dxLM>-7</R_dxLM>
<R_dyLM>-24</R_dyLM>
<R_xC>202.0</R_xC>
<R_yC>167.0</R_yC>
<R_merX1>201</R_merX1>
<R_merX2>203</R_merX2>
<R_merY1>164</R_merY1>
<R_merY2>169</R_merY2>
<S_spotNbr>759</S_spotNbr>
<S_dxLM>-2</S_dxLM>
<S_dyLM>-24</S_dyLM>
<S_xC>226.0</S_xC>
<S_yC>167.0</S_yC>
<S_merX1>224</S_merX1>
<S_merX2>229</S_merX2>
<S_merY1>152</S_merY1>
<S_merY2>158</S_merY2>
<PairingCode>A</PairingCode>
<DP>5.4</DP>
<DL>25.0</DL>
<R_area>11</R_area>
<S_area>25</S_area>
<R_dens>1.470</R_dens>
<S_dens>8.352</S_dens>
<R_dPrime>0.961</R_dPrime>
<S_dPrime>3.515</S_dPrime>
<R_volume>1.125</R_volume>
<S_volume>6.484</S_volume>
<R_MaxDens>0.186</R_MaxDens>
<S_MaxDens>0.480</S_MaxDens>
<R_MinDens>0.043</R_MinDens>
<S_MinDens>0.225</S_MinDens>
<R_MeanBkgDens>0.046</R_MeanBkgDens>
<S_MeanBkgDens>0.193</S_MeanBkgDens>
<R_stdDev_X>0.704</R_stdDev_X>
<R_stdDev_Y>1.209</R_stdDev_Y>
<S_stdDev_X>1.292</S_stdDev_X>
<S_stdDev_Y>1.473</S_stdDev_Y>
</Paired_Spot>
. . .
<Paired_Spot>
<LandmarkSet>V</LandmarkSet>
<R_spotNbr>0</R_spotNbr>
<R_dxLM>0</R_dxLM>
<R_dyLM>0</R_dyLM>
<R_xC>0.0</R_xC>
<R_yC>0.0</R_yC>
<R_merX1>0</R_merX1>
<R_merX2>0</R_merX2>
<R_merY1>0</R_merY1>
<R_merY2>0</R_merY2>
<S_spotNbr>1422</S_spotNbr>
<S_dxLM>159</S_dxLM>
<S_dyLM>0</S_dyLM>
<S_xC>446.0</S_xC>
<S_yC>0.0</S_yC>
<S_merX1>441</S_merX1>
<S_merX2>448</S_merX2>
<S_merY1>493</S_merY1>
<S_merY2>499</S_merY2>
<PairingCode>U</PairingCode>
<DP>166.5</DP>
<DL>166.5</DL>
<R_area>0</R_area>
<S_area>33</S_area>
<R_dens>0.000</R_dens>
<S_dens>0.630</S_dens>
<R_dPrime>0.000</R_dPrime>
<S_dPrime>0.395</S_dPrime>
<R_volume>0.000</R_volume>
<S_volume>0.601</S_volume>
<R_MaxDens>0.000</R_MaxDens>
<S_MaxDens>0.041</S_MaxDens>
<R_MinDens>0.000</R_MinDens>
<S_MinDens>0.000</S_MinDens>
<R_MeanBkgDens>0.000</R_MeanBkgDens>
<S_MeanBkgDens>0.007</S_MeanBkgDens>
<R_stdDev_X>0.000</R_stdDev_X>
<R_stdDev_Y>0.000</R_stdDev_Y>
<S_stdDev_X>1.512</S_stdDev_X>
<S_stdDev_Y>1.365</S_stdDev_Y>
</Paired_Spot>
<Global_Spot_pairing_statistics>
<NbrRsampleSpotsInLMS>738</NbrRsampleSpotsInLMS>
<NbrSampleSpotsInLMS>1427</NbrSampleSpotsInLMS>
<Landmark_set_sizes>
<Landmark>
<Landmark_name>A</Landmark_name>
<Landmark_nbr>1</Landmark_nbr>
<Nbr_Rsample_spots>12</Nbr_Rsample_spots>
<Nbr_Sample_spots>18</Nbr_Sample_spots>
</Landmark>
<Landmark>
<Landmark_name>B</Landmark_name>
<Landmark_nbr>2</Landmark_nbr>
<Nbr_Rsample_spots>34</Nbr_Rsample_spots>
<Nbr_Sample_spots>77</Nbr_Sample_spots>
</Landmark>
. . .
<Landmark>
<Landmark_name>V</Landmark_name>
<Landmark_nbr>22</Landmark_nbr>
<Nbr_Rsample_spots>39</Nbr_Rsample_spots>
<Nbr_Sample_spots>93</Nbr_Sample_spots>
</Landmark>
</Landmark_set_sizes>
<InitialpairingStats>
<Nbr_US_spotsPri>608</Nbr_US_spotsPri>
<Nbr_SP_spotsPri>230</Nbr_SP_spotsPri>
<Nbr_PP_spotsPri>764</Nbr_PP_spotsPri>
<Nbr_AP_spotsPri>563</Nbr_AP_spotsPri>
<Nbr_CP_spotsPri>0</Nbr_CP_spotsPri>
<Nbr_EP_spotsPri>0</Nbr_EP_spotsPri>
</InitialpairingStats>
<SecondarypairingStats>
<Nbr_US_spotsSec>594</Nbr_US_spotsSec>
<Nbr_SP_spotsSec>230</Nbr_SP_spotsSec>
<Nbr_PP_spotsSec>810</Nbr_PP_spotsSec>
<Nbr_AP_spotsSec>531</Nbr_AP_spotsSec>
<Nbr_CP_spotsSec>0</Nbr_CP_spotsSec>
<Nbr_EP_spotsSec>0</Nbr_EP_spotsSec>
</SecondarypairingStats>
<Primary_SP_PP_pairRate>67.3</Primary_SP_PP_pairRate>
<Secondary_SP_PP_pairRate>70.5</Secondary_SP_PP_pairRate>
<meanDP_SP_PP>10.9</meanDP_SP_PP>
<meanDPprime_SP_PP>17.6</meanDPprime_SP_PP>
</Global_Spot_pairing_statistics>
</SpotList_Pairing>
8.4 Examples - part of Sample Pairs File GELLAB-II format
This is part of a Sample Pairs File to illustrate the type of data
available as output. This used the default -spfFormat:X option.
CmpSpots V.0.1.0-pre-Alpha - $Date$ - $Revision$ (Open2Dprot)
Input Rsample file: gel-HM-019.gif
Input Sample file: gel-HM-071.gif
Output Sample Pairs File: demo\xml\gel-HM-071.spf
Spot lists parameters and statistics
------------------------------------
Rsample Window [14:475,74:509] (pixels) [rows,cols]=[512,512] PixelSizeMicrons=0.00
NbrSpots=3072 NbrSpotsPrime=1717 NbrSpotsOmitted= 1355
TotSampleDensity=80411.00 TotSampleDensityPrime=26247.40 TotOmittedDensity=121.00
TotSampleArea=29974 TotSampleAreaPrime=55093 TotSampleAreaOmitted=22282
Sample Window [6:450,68:503] (pixels) [rows,cols]=[512,512] PixelSizeMicrons=0.00
NbrSpots=1484 NbrSpotsPrime=864 NbrSpotsOmitted= 620
TotSampleDensity=44242.00 TotSampleDensityPrime=16980.60 TotOmittedDensity=50.80
TotSampleArea=21961 TotSampleAreaPrime=33078 TotSampleAreaOmitted=11164
Sample pairs threshold sizing parameters
----------------------------------------
Threshold Sure-Pair (SP) sizing limit, thrSP: 5.0 (pixels)
Threshold Possible-Pair (PP) sizing limit, thrPP: 10.0 (pixels)
Number of alternate landmark sets to check: 1
Switches: -noaccessionFile:accession.xml -noaltLandmarks:1 -nocommutativeLMS
-nocsamplePrimeData -nodebugBits:0,0 -nodefault -demo
-nodtd -histGUI -inputFormat:X,X,X -nolandmarkFile:landmark.xml
-latchLandmarkSpots:6.0 -nopercentDensity -noprojDir:demo\
-nopropertiesFile:CmpSpots.properties -rsample:gel-HM-019
-sample:gel-HM-071 -spfFormat:G -secondaryPairing -nossfCW:0,0,0,0
-thrSP:5.00 -thrPP:10.00 -notimer -nounCorrectedDensity
-noupdate:program
#A R:209 d(xy)LM1[-2,-26] (xy)C1[205.0,164.0] MER1[202:208,162:167] S:178 d(xy)LM2[-2,-28] (xy)C2[225.0,148.0] MER2[222:228,147:151]P dP=2.0,dL=28.131area1 22area2 9.190D1gray 5.495D1gray 7.870D'1 1.238D'2 7.756V'1 4.037V'2 0.540Maxd1 0.325Maxd2 0.070MinD1 0.122MinD2 1.348sX1 1.517sX2 1.503sY1 1.155sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.043MnBk1 0.193MnBk2
#A R:197 d(xy)LM1[6,-20] (xy)C1[233.0,156.0] MER1[209:213,164:168] S:219 d(xy)LM2[4,-24] (xy)C2[211.0,166.0] MER2[230:236,153:160]A dP=4.5,dL=24.343area1 15area2 14.598D1gray 5.336D1gray 7.120D'1 4.780D'2 11.566V'1 3.308V'2 0.540Maxd1 0.376Maxd2 0.174MinD1 0.275MinD2 1.685sX1 1.244sX2 1.792sY1 0.997sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.174MnBk1 0.037MnBk2
#A R:233 d(xy)LM1[-1,-19] (xy)C1[206.0,171.0] MER1[203:208,168:175] S:196 d(xy)LM2[-1,-21] (xy)C2[226.0,155.0] MER2[224:229,152:158]P dP=2.0,dL=21.034area1 25area2 10.213D1gray 8.352D1gray 8.702D'1 3.515D'2 8.683V'1 6.484V'2 0.480Maxd1 0.480Maxd2 0.122MinD1 0.225MinD2 1.433sX1 1.292sX2 1.779sY1 1.473sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.044MnBk1 0.193MnBk2
#A R:234 d(xy)LM1[6,-18] (xy)C1[213.0,172.0] MER1[209:219,168:176] S:197 d(xy)LM2[6,-20] (xy)C2[233.0,156.0] MER2[230:236,153:160]P dP=2.0,dL=20.946area1 43area2 18.568D1gray 14.598D1gray 16.779D'1 7.120D'2 17.748V'1 11.566V'2 0.540Maxd1 0.540Maxd2 0.325MinD1 0.174MinD2 2.545sX1 1.685sX2 1.822sY1 1.792sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.039MnBk1 0.174MnBk2
#A R:252 d(xy)LM1[9,-13] (xy)C1[216.0,177.0] MER1[213:219,175:178] S:217 d(xy)LM2[7,-12] (xy)C2[234.0,164.0] MER2[231:237,161:167]P dP=2.2,dL=15.821area1 37area2 15.298D1gray 9.663D1gray 14.365D'1 0.876D'2 12.723V'1 7.212V'2 1.164Maxd1 0.376Maxd2 0.480MinD1 0.122MinD2 1.570sX1 1.650sX2 0.981sY1 1.639sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.044MnBk1 0.237MnBk2
. . .
#A R:278 d(xy)LM1[6,13] (xy)C1[233.0,189.0] MER1[212:221,203:213] S:1111 d(xy)LM2[9,18] (xy)C2[216.0,208.0] MER2[229:238,185:193]A dP=5.8,dL=20.161area1 64area2 56.456D1gray 15.077D1gray 34.708D'1 10.568D'2 50.597V'1 12.561V'2 1.700Maxd1 0.396Maxd2 0.174MinD1 0.077MinD2 2.089sX1 1.835sX2 2.009sY1 2.441sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.357MnBk1 0.070MnBk2
#A R:223 d(xy)LM1[0,-12] (xy)C1[227.0,164.0] MER1[224:229,160:165] S:223 d(xy)LM2[20,-26] (xy)C2[227.0,164.0] MER2[224:229,160:165]A dP=5.8,dL=12.016area1 16area2 6.412D1gray 6.412D1gray 2.112D'1 2.112D'2 7.021V'1 7.021V'2 0.540Maxd1 0.540Maxd2 0.225MinD1 0.225MinD2 1.435sX1 1.435sX2 1.278sY1 1.278sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.269MnBk1 0.269MnBk2
#A R:266 d(xy)LM1[23,4] (xy)C1[250.0,180.0] MER1[246:252,177:183] S:266 d(xy)LM2[43,-10] (xy)C2[250.0,180.0] MER2[246:252,177:183]A dP=7.8,dL=23.338area1 38area2 32.619D1gray 32.619D1gray 21.457D'1 21.457D'2 27.195V'1 27.195V'2 1.392Maxd1 1.392Maxd2 0.480MinD1 0.480MinD2 1.636sX1 1.636sX2 1.685sY1 1.685sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.294MnBk1 0.294MnBk2
#B R:1 d(xy)LM1[0,-82] (xy)C1[208.0,71.0] MER1[204:210,69:73] S:0 d(xy)LM2[31,0] (xy)C2[0.0,0.0] MER2[0:0,0:0]U dL=10.0,dL=87.324area1 0area2 0.971D1gray 0.000D1gray 0.971D'1 0.000D'2 0.995V'1 0.000V'2 0.070Maxd1 0.000Maxd2 0.027MinD1 0.000MinD2 1.572sX1 0.000sX2 1.283sY1 0.000sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.000MnBk1 0.000MnBk2
#B R:26 d(xy)LM1[0,-77] (xy)C1[158.0,76.0] MER1[155:162,73:78] S:0 d(xy)LM2[-17,0] (xy)C2[0.0,0.0] MER2[0:0,0:0]U dL=10.4,dL=79.634area1 0area2 14.270D1gray 0.000D1gray 13.703D'1 0.000D'2 11.852V'1 0.000V'2 0.915Maxd1 0.000Maxd2 0.027MinD1 0.000MinD2 1.840sX1 0.000sX2 0.993sY1 0.000sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.017MnBk1 0.000MnBk2
#B R:27 d(xy)LM1[0,-77] (xy)C1[165.0,76.0] MER1[163:167,73:78] S:0 d(xy)LM2[-17,0] (xy)C2[0.0,0.0] MER2[0:0,0:0]U dL=10.8,dL=78.119area1 0area2 6.880D1gray 0.000D1gray 6.598D'1 0.000D'2 5.253V'1 0.000V'2 0.667Maxd1 0.000Maxd2 0.027MinD1 0.000MinD2 1.187sX1 0.000sX2 0.936sY1 0.000sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.015MnBk1 0.000MnBk2
#B R:28 d(xy)LM1[0,-77] (xy)C1[170.0,76.0] MER1[168:171,73:78] S:0 d(xy)LM2[-17,0] (xy)C2[0.0,0.0] MER2[0:0,0:0]U dL=13.5,dL=77.416area1 0area2 2.850D1gray 0.000D1gray 2.613D'1 0.000D'2 2.701V'1 0.000V'2 0.428Maxd1 0.000Maxd2 0.027MinD1 0.000MinD2 0.925sX1 0.000sX2 0.962sY1 0.000sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.015MnBk1 0.000MnBk2
. . .
#U R:0 d(xy)LM1[0,0] (xy)C1[0.0,0.0] MER1[0:0,0:0] S:862 d(xy)LM2[20,65] (xy)C2[256.0,498.0] MER2[252:259,494:501]U dL=68.0,dL=68.00area1 52area2 0.000D1gray 36.111D1gray 34.904D'1 34.904D'2 30.105V'1 30.105V'2 0.000Maxd1 1.392Maxd2 0.000MinD1 0.070MinD2 0.000sX1 1.777sX2 0.000sY1 1.716sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.000MnBk1 0.023MnBk2
#U R:0 d(xy)LM1[0,0] (xy)C1[0.0,0.0] MER1[0:0,0:0] S:863 d(xy)LM2[-68,67] (xy)C2[168.0,500.0] MER2[164:171,497:502]U dL=95.5,dL=95.50area1 38area2 0.000D1gray 10.867D1gray 9.917D'1 9.917D'2 9.453V'1 9.453V'2 0.000Maxd1 0.540Maxd2 0.000MinD1 0.070MinD2 0.000sX1 1.789sX2 0.000sY1 1.380sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.000MnBk1 0.025MnBk2
#U R:0 d(xy)LM1[0,0] (xy)C1[0.0,0.0] MER1[0:0,0:0] S:864 d(xy)LM2[-86,68] (xy)C2[150.0,501.0] MER2[147:153,499:502]U dL=99.0,dL=109.60area1 19area2 0.000D1gray 0.975D1gray 0.534D'1 0.534D'2 1.143V'1 1.143V'2 0.000Maxd1 0.122Maxd2 0.000MinD1 0.027MinD2 0.000sX1 1.457sX2 0.000sY1 0.909sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.000MnBk1 0.023MnBk2
#V R:1510 d(xy)LM1[-46,13] (xy)C1[241.0,431.0] MER1[238:244,427:435] S:757 d(xy)LM2[-40,8] (xy)C2[276.0,400.0] MER2[272:280,396:404]P dP=7.8,dL=47.846area1 50area2 11.915D1gray 36.901D1gray 10.297D'1 34.401D'2 9.979V'1 32.682V'2 0.500Maxd1 1.392Maxd2 0.043MinD1 0.174MinD2 1.649sX1 1.809sX2 1.707sY1 1.830sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.035MnBk1 0.050MnBk2
#V R:658 d(xy)LM1[0,-63] (xy)C1[322.0,355.0] MER1[318:326,351:360] S:0 d(xy)LM2[29,0] (xy)C2[0.0,0.0] MER2[0:0,0:0]U dL=13.4,dL=72.157area1 0area2 42.779D1gray 0.000D1gray 41.829D'1 0.000D'2 38.131V'1 0.000V'2 1.700Maxd1 0.000Maxd2 0.070MinD1 0.000MinD2 1.887sX1 0.000sX2 1.677sY1 0.000sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.017MnBk1 0.000MnBk2
#V R:671 d(xy)LM1[0,-56] (xy)C1[371.0,362.0] MER1[368:376,359:365] S:0 d(xy)LM2[55,0] (xy)C2[0.0,0.0] MER2[0:0,0:0]U dL=38.9,dL=101.043area1 0area2 1.887D1gray 0.000D1gray 1.569D'1 0.000D'2 2.180V'1 0.000V'2 0.122Maxd1 0.000Maxd2 0.000MinD1 0.000MinD2 1.740sX1 0.000sX2 1.452sY1 0.000sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.007MnBk1 0.000MnBk2
#V R:660 d(xy)LM1[37,-47] (xy)C1[353.0,345.0] MER1[326:334,359:366] S:674 d(xy)LM2[43,-55] (xy)C2[330.0,363.0] MER2[351:356,341:349]A dP=10.0,dL=69.828area1 49area2 1.416D1gray 20.034D1gray 1.016D'1 19.490D'2 1.828V'1 16.801V'2 0.122Maxd1 0.723Maxd2 0.027MinD1 0.122MinD2 1.398sX1 1.922sX2 1.514sY1 1.705sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.014MnBk1 0.011MnBk2
. . .
#V R:0 d(xy)LM1[0,0] (xy)C1[0.0,0.0] MER1[0:0,0:0] S:827 d(xy)LM2[65,60] (xy)C2[381.0,452.0] MER2[378:384,449:455]U dL=99.0,dL=101.40area1 27area2 0.000D1gray 0.484D1gray 0.339D'1 0.339D'2 0.474V'1 0.474V'2 0.000Maxd1 0.041Maxd2 0.000MinD1 0.000MinD2 0.000sX1 1.437sX2 0.000sY1 1.134sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.000MnBk1 0.005MnBk2
#V R:0 d(xy)LM1[0,0] (xy)C1[0.0,0.0] MER1[0:0,0:0] S:836 d(xy)LM2[5,70] (xy)C2[321.0,462.0] MER2[317:324,459:465]U dL=70.2,dL=70.20area1 40area2 0.000D1gray 3.944D1gray 3.444D'1 3.444D'2 3.371V'1 3.371V'2 0.000Maxd1 0.174Maxd2 0.000MinyD1 0.041MinD2 0.000sX1 1.943sX2 0.000sY1 1.408sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.000MnBk1 0.013MnBk2
#V R:0 d(xy)LM1[0,0] (xy)C1[0.0,0.0] MER1[0:0,0:0] S:855 d(xy)LM2[15,96] (xy)C2[331.0,488.0] MER2[327:333,486:491]U dL=97.2,dL=97.20area1 22area2 0.000D1gray 0.580D1gray 0.462D'1 0.462D'2 0.480V'1 0.480V'2 0.000Maxd1 0.041Maxd2 0.000MinD1 0.000MinD2 0.000sX1 1.330sX2 0.000sY1 1.240sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.000MnBk1 0.005MnBk2
#V R:0 d(xy)LM1[0,0] (xy)C1[0.0,0.0] MER1[0:0,0:0] S:857 d(xy)LM2[37,103] (xy)C2[353.0,495.0] MER2[349:357,491:498]U dL=99.0,dL=109.40area1 47area2 0.000D1gray 1.691D1gray 1.439D'1 1.439D'2 1.617V'1 1.617V'2 0.000Maxd1 0.070Maxd2 0.000MinD1 0.000MinD2 0.000sX1 1.849sX2 0.000sY1 1.774sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.000MnBk1 0.005MnBk2
#V R:0 d(xy)LM1[0,0] (xy)C1[0.0,0.0] MER1[0:0,0:0] S:859 d(xy)LM2[130,104] (xy)C2[446.0,496.0] MER2[441:448,493:499]U dL=99.0,dL=177.10area1 33area2 0.000D1gray 0.630D1gray 0.395D'1 0.395D'2 0.601V'1 0.601V'2 0.000Maxd1 0.041Maxd2 0.000MinD1 0.000MinD2 0.000sX1 1.512sX2 0.000sY1 1.365sY2 0.00sdMX,0.00sdMY,0.00sdMD0.00sdMA,0#G,0.000MnBk1 0.007MnBk2
Summary: of paired-spot statistics
----------------------------------
Rsample has 1564, Sample 864 spots in all landmark sets.
After Initial pairing:
US 107
SP 312
PP 882
AP 671
CP 0
EP 0
0.5(SP+PP)/(|G1| MIN |G2|)=69.10%
After secondary pairing:
US 95
SP 312
PP 918
AP 647
CP 0
EP 0
After secondary pairing: 0.5(SP+PP)/(|G1| MIN |G2|)=71.2%
mean dP(SP+PP)=1.88, mean dP'((|G1|+|G2|)/(SP+PP))=0.63
List of image files and generated files
---------------------------------------
Input Rsample pix file [demo\ppx\gel-HM-019.gif]
Input Sample pix file [demo\ppx\gel-HM-071.gif]
Input Rsample SSF file [demo\xml\gel-HM-019.xml]
Input Sample SSF file [demo\xml\gel-HM-071.xml]
Output SPF file [demo\xml\gel-HM-071.spf]
FINISHED! The Sample Paired-spotlist File (SPF), is demo\xml\gel-HM-071.spf
Run time =0:0:1 (H:M:S) or 1.6 seconds
8.5 Examples - using command line processing for batch
It is possible to run the CmpSpots from the command line in your
operating system. We give two examples doing this. The first example
shows a script for the Microsoft Windows batch (.bat) file demo-CmpSpots.bat file (available on the Files
Mirror. The second example
shows the same commands in a shell script for a Unix operating system
(Linux, MacOS, Solaris, etc.) that you can download with demo-CmpSpots.sh.
8.5.1 Examples - batch processing under Microsoft Windows
REM File: demo-CmpSpots.bat - pair spots to the Reference sample for a list of samples
REM This example assumes that all .jar files listed below and demo/ directory are
REM in the current directory. Modify for other situations.
REM
REM The JDK should be installed and version 1.4 or later is required.
REM You can download the latest JDK from http://java.sun.com/
REM
REM The files needed are listed below:
REM JAR files required and mentioned in manifest:
REM JAR files:
REM xml-apis.jar xercesImpl.jar jai_codec.jar jai_core.jar O2Plib.jar
REM
REM demo image files:
REM demo/ppx/gel-HM-019 (Reference sample)
REM demo/ppx/gel-HM-071
REM demo/ppx/gel-HM-087
REM demo/ppx/gel-HM-096
REM Accession database file is in:
REM demo/xml/accession.xml
REM Landmark database file is in:
REM demo/xml/landmark.xml
REM Sample Spot-list files (SSF) are in:
REM demo/xml/gel-HM-019-SSF.xml
REM demo/xml/gel-HM-071-SSF.xml
REM demo/xml/gel-HM-087-SSF.xml
REM demo/xml/gel-HM-096-SSF.xml
REM Generated Sample Paired-spot-list Files (SPF) are saved in:
REM demo/xml/
REM Generated images are saved in:
REM demo/tmp/
REM
REM P. Lemkin $Date$
echo "demo-CmpSpots.bat"
pwd
date /T
java -Xmx256M -jar CmpSpots.jar -default -project:demo\ -sample:gel-HM-071 -rsample:gel-HM-019
java -Xmx256M -jar CmpSpots.jar -default -project:demo\ -sample:gel-HM-087 -rsample:gel-HM-019
java -Xmx256M -jar CmpSpots.jar -default -project:demo\ -sample:gel-HM-096 -rsample:gel-HM-019
echo "-- Finished pairing the samples ---"
date /T
8.5.1.1 Examples - batch processing with GUI under Microsoft Windows
REM File: demo-CmpSpots-GUI.bat - pair spots to the Reference sample for a sample using the GUI
REM This example assumes that all .jar files listed below and demo/ directory are
REM in the current directory. Modify for other situations.
REM
REM The JDK should be installed and version 1.4 or later is required.
REM You can download the latest JDK from http://java.sun.com/
REM
REM The files needed are listed below:
REM JAR files required and mentioned in manifest:
REM xml-apis.jar xercesImpl.jar jai_codec.jar jai_core.jar O2Plib.jar
REM
REM demo image files:
REM demo/ppx/gel-HM-019 (Reference sample)
REM demo/ppx/gel-HM-071
REM demo/ppx/gel-HM-087
REM demo/ppx/gel-HM-096
REM Accession database file is in:
REM demo/xml/accession.xml
REM Landmark database file is in:
REM demo/xml/landmark.xml
REM Sample Spot-list files (SSF) are in:
REM demo/xml/gel-HM-019-SSF.xml
REM demo/xml/gel-HM-071-SSF.xml
REM demo/xml/gel-HM-087-SSF.xml
REM demo/xml/gel-HM-096-SSF.xml
REM Generated Sample Paired-spot-list Files (SPF) are saved in::
REM demo/xml/
REM Generated images are saved in:
REM demo/tmp/
REM
REM P. Lemkin $Date$
echo "demo-CmpSpots-GUI.bat"
pwd
date /T
java -Xmx256M -jar CmpSpots.jar -demo -project:demo\ -sample:gel-HM-071 -rsample:gel-HM-019
echo "-- Finished pairing the sample ---"
date /T
8.5.2 Examples - batch processing under Unix
Because java is relatively operating system independent, the same java
command lines are used with the "\" changed to "/" from Windows to
Unix file path convention.
#!/bin/sh
# File: demo-CmpSpots.sh - Unix script to pair spots to the
# Reference sample for a list of samples
# This example assumes that all .jar files listed below and demo/ directory are
# in the current directory. Modify for other situations.
#
# The JDK should be installed and version 1.4 or later is required.
# You can download the latest JDK from http://java.sun.com/
#
# The files needed are listed below:
# JAR files required and mentioned in manifest:
# xml-apis.jar xercesImpl.jar jai_codec.jar jai_core.jar O2Plib.jar
#
# demo sample image files:
# demo/ppx/gel-HM-019 (Reference sample)
# demo/ppx/gel-HM-071
# demo/ppx/gel-HM-087
# demo/ppx/gel-HM-096
# Accession database file is in:
# demo/xml/accession.xml
# Landmark database file is in:
# demo/xml/landmark.xml
# Sample Spot-list files (SSF) are in:
# demo/xml/gel-HM-019-SSF.xml
# demo/xml/gel-HM-071-SSF.xml
# demo/xml/gel-HM-087-SSF.xml
# demo/xml/gel-HM-096-SSF.xml:
# Generated Sample Paired-spot-list Files (SPF) are saved in:
# demo/xml/
# Generated images are saved in:
# demo/tmp/
#
# P. Lemkin $Date$
echo "demo-CmpSpots.sh"
pwd
date
java -Xmx256M -jar CmpSpots.jar -default -project:demo/ -sample:gel-HM-071 -rsample:gel-HM-019
java -Xmx256M -jar CmpSpots.jar -default -project:demo/ -sample:gel-HM-087 -rsample:gel-HM-019
java -Xmx256M -jar CmpSpots.jar -default -project:demo/ -sample:gel-HM-096 -rsample:gel-HM-019
echo "-- Finished pairing the samples ---"
date
8.5.2.1 Examples - batch processing with GUI under Unix
#!/bin/sh
# File: demo-CmpSpots-GUI.sh - Unix script to pair spots to the
# Reference sample for a sample using the GUI.
# This example assumes that all .jar files listed below and demo/ directory are
# in the current directory. Modify for other situations.
#
# The JDK should be installed and version 1.4 or later is required.
# You can download the latest JDK from http://java.sun.com/
#
# The files needed are listed below:
# JAR files required and mentioned in manifest:
# xml-apis.jar xercesImpl.jar jai_codec.jar jai_core.jar O2Plib.jar
#
# demo sample image files:
# demo/ppx/gel-HM-019 (Reference sample)
# demo/ppx/gel-HM-071
# Accession database file is in:
# demo/xml/accession.xml
# Landmark database file is in:
# demo/xml/landmark.xml
# Sample Spot-list files (SSF) are in:
# demo/xml/gel-HM-019-SSF.xml
# demo/xml/gel-HM-071-SSF.xml
# demo/xml/gel-HM-087-SSF.xml
# demo/xml/gel-HM-096-SSF.xml:
# Generated Sample Paired-spot-list Files (SPF) are saved in:
# demo/xml/
# Generated images are saved in:
# demo/tmp/
#
# P. Lemkin $Date$
echo "demo-CmpSpots-GUI.sh"
pwd
date
java -Xmx256M -jar CmpSpots.jar -demo -gui -project:demo/ -sample:gel-HM-071 -rsample:gel-HM-019
echo "-- Finished pairing the samples ---"
date
9. CmpSpots References
These papers (a subset of the GELLAB-II papers),
reference the GELLAB-II spot pairing program. The Open2Dprot
Java-language CmpSpots program was derived from the GELLAB-II
C-language program as well as from code from the MAExplorer and Flicker projects.
New Java code was added as well. Although CmpSpots has been enhanced
in many ways, the basic algorithm is similar so these papers may be
useful for more details on the algorithm.
Contact us
CmpSpots is a contributed program available at
open2dprot.sourceforge.net/CmpSpots
Powered by
Revised: 05/19/2006