Landmark Reference Manual

Landmark Reference Manual
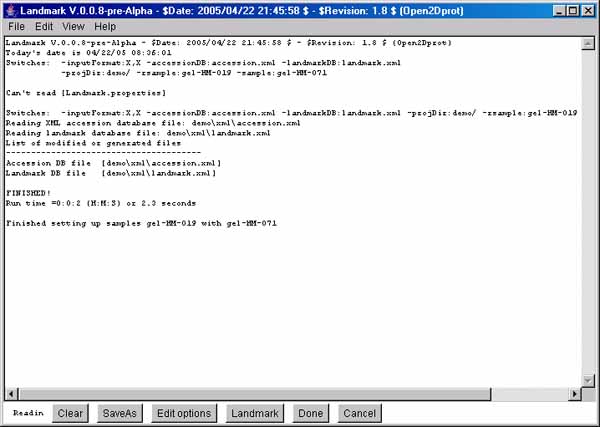
Figure 1. Screen view of the Landmark program Report Window interface
This screen shot shows the initial Landmark Report
Window graphical user interface available after the program
starts. There are a number of pull-down menus (File, Edit, View, and Help) as well
as pushbuttons at the bottom of the window. Edit Options lets you change the
startup options. The Landmark button
causes the interactive Landmark Editor window to pop up. When you are
finished editing a landmark set, pressing the Done button will
save the data set in the landmark database file. You can also do these
operations from the menus.
1. Introduction
Landmark is an open source Java 2D sample Landmarking program for
creating landmark sets for samples paired with a reference sample
(Rsample) into the Open2Dprot database.
It is part of the Open2Dprot project (
http://open2dprot.sourceforge.net/).
Landmark is similar to and derived from an earlier landmark program in
the GELLAB-II 2D gel spot pairing program landmark described in
the GELLAB-II reference manual. Landmark has been generalized to other
types of samples besides 2D gels. While the original program was
written in C, Landmark is written in Java, uses XML input and output
files and has an Java-based graphical user interface. This initial
open-source Landmark program code could be used as the basis for more
advance spot pairing methods. Currently the Landmark file is
landmark.xml and is stored in the project
"project"/xml/ directory.
The program is run interactively (-gui). After the spot pairing is finished, the user has the option of interactively examining the paired spot data overlayed on the original sample images. The user may also modify the input switch options and save the new options in a "Landmark.properties" file in the current project directory when they exit so that the last used options may be used as the default switch options in subsequent running of Landmark.
The application looks up the sample in the accession database (in
xml/accession.xml or as specified using the -accessionFile switch) and
gets additional information about the sample. The Open2Dprot
http://open2dprot.sourceforge.net/Accession pipeline module is
used for entering samples into the accession database.
The images associated with the samples can be used with the landmark
editor. These images may be in TIFF (.tif, .tiff), JPEG (.jpg), GIF
(.gif), or PPX (.ppx GELLAB-II) format. TIFF images may be
8-bits/pixel through 16-bits/pixel, whereas JPEG, GIF, and PPX are
8-bit images. Gray values in the image files have black as 0. This is
mapped to 0 for white and the maximum pixel value for black.
The input sample image files, if any, are kept in the
user-project-directory/ppx/ sub-directory. This database
directory structure is consistent with and is used by the other
Open2Dprot analysis pipeline programs.
Normally the project directory is setup prior to running Landmark and
it expects the data to be in the following directories. You can also
use the -projDir:user-project-directory switch to specify a
(possibly new) project directory.
If you are using the Windows Landmark.exe file or clicking on the
Landmark.jar file, you can't change the default startup memory.
If you are using the Landmark.jar in a script using the java interpreter
as in the following example which uses the -Xmx256M
(specifying using 256Mbytes at startup). Change 256 to a larger size
if you want to increase startup memory.
All logged output is sent to the report window in a scrollable text
window that may be saved or used for cut and paste operations. A set
of command buttons at the bottom of the window are replicates of
commands in the menus, but are easier to access. They include the
following functions:
The documentation is kept on the Internet at
http://open2dprot.sourceforge.net/Landmark. Normally, these help
commands should pop up a Web browser that directly points to the
Landmark Web page. If your browser is not configured correctly, it
may not be able to be launched directly from the Landmark
program. Instead, just go to the Web site with your Web browser and
look up the information there.
The menu bar a the top of the Report Window (see Figure 1) contains
four menus. There are also some command buttons at the bottom of the
window for convenience that are also menu commands.
There are several ways to run the program. On Windows, you can start
Landmark by clicking on the startup icon shown in Figure 6 below.
For Unix systems including MacOS-X, you can start Landmark from
the command line by running the Landmark.jar file. If your computer is setup
to execute jar files, just type the jar file. In both systems, you can specify
additional command line arguments in Windows .bat and unix .sh scripts (see
demo examples below.
An Internet connection is required to download the program from the
Open2Dprot Landmark Web site. New versions of the program and
associated demo data will become available on this Web site and can be
uploaded to your computer using the various
(File | Update | ...) menu commands. If you have obtained the
installer software that someone else downloaded and gave to you, then
you do not need the Internet connection to install the program. We
currently distribute Landmark so that it uses up to 256Mb. See discussion
on increasing memory.
You can these images in the list below or view all of the screen shots in a
single Web page.
The data for these scripts is in the demo/ directory available with the
installation which also includes the scripts.
Alternatively, you can download the demo data from the Files Mirror as
Demo.Z.
The installation packages are available from the
Files mirror
under the Landmark releases.
Project directory structure for Open2Dprot and Landmark
All Open2Dprot programs assume a project directory structure. This
must exist for the program to proceed. You can either create the
structure prior to running any of the programs or you can create it on
the fly using the -projDir:user-project-directory. It will
lookup and/or create the following sub-directories inside of
theuser-project-directory.
batch/ directory holding temporary batch files - [NOT USED by Landmark]
cache/ directory holding temporary CSD cache files - [NOT USED by Landmark]
ppx/ directory holding your original gel input files
rdbms/ directory holding CSD database RDBMS files - [NOT USED by Landmark]
tmp/ directory holding generated sample image files
xml/ directory that holds Accession DB, landmark DB,
SSF sample spot-list files, and SPF samples paired-spot-list files
The use of these directories is discussed in the rest of this
document.Landmark database file
The sample and reference samples to be paired is specified by its
sample name using the -sample and -rsample command line switches. It
can also be specified at run time using the (File | Open Rsample |
...) and (File | Open Sample | ...) menu commands.2. Running Landmark and specifying parameter options via the
command line
The program is run interactively (-gui) with a graphical user
interface (GUI). The user may also modify the input switch options
and save the new options in a "Landmark.properties" file in the
current project directory so that it may be used as the default switch
options in subsequent running of the program.
[Status: the Landmark.properties file is not enabled.]
All options including the input reference sample and other sample to
be paired are specified via GNU/Unix style switches on the command
line (-switch{optional ':parameters'} and its negation as
-noswitch). However, if GUI mode is used, you can interactively
specify the switches and their options. It is assigned previously by
software that generated the SSF spot lists.
Local Folders and files created and used by Landmark
When Landmark is first started, it will check for the following folders
and files in the installation directory and create them if they can
not be found.
Landmark command-line arguments switch usage
The command line arguements usage is:
Landmark -rsample:Reference-sample -sample:sample [< optional switches >]
The complete list of switches is given
later in this manual and as well as some examples of typical sets of switches. The
user defined default switches may be specified as a resource string
'Landmark.properties' file saved in the project directory. For
example:
Landmark -rsample:gel-HM-19 -sample:gel-HM-071 -project:demo/ -gui
Options wizard window for setting the command line switches
If you invoke the Edit options button in the Report window (or
from the Edit menu), it will popup an options wizard shown in Figure 2
to let you set or change the switch options and then to save these as
the new default switch options. The default is saved in the
Landmark.properties file when you exit program.

Figure 2. Screen view of the popup options wizard window for
setting the command line switches, parameter and specifying input
samples to be paired.
All of the switches are available in the
scrollable window. Switches are checked if they are enabled and if the
switch requires a value, the current value is shown in the data entry
window to its right. On the right there may be several threshold
sliders for the upper sizing values for several parameters. In the
middle, are several Browse buttons to use for specifying a different
samples (-rsample: and -sample:), and directories.
Clicking on any switch will show a short help message associated with
that switch at the top of the window. Pressing the Set new
default button will pass the new options values back to
Landmark. Note: for this to take effect, you must exit and then
restart Landmark. Then to use them, press the Pair spots
button in the main Report Window.
Updating Landmark from the Open2Dprot Web server using -update switch
As new versions of Landmark are developed and put on the Web server,
a more efficient way of updating your version is to use the -update
commands. There are four options:
-update:program to update the program jar file
-update:demo to update the demonstration files
-update:doc to update the documentation files
-update:all to update all of the above
After updating the program, it should be exited and restarted for the
new program to take effect. Increasing the allowable memory used by Landmark
If you are working with very large images that require a lot of memory,
you might want to increase the memory available at startup.
java -Xmx256M -jar Landmark.jar {additional command line args}
3. Command and Report Window - the command center
Landmark is designed to be used with the graphical user interface
(GUI) which creates a Report
Window that captures a report of the spot pairing output as well
as additional output directed to it by the user. There are a set of pull-down menus as well as a set of buttons for
often used functions.
4. Pull-down menus in the Landmark Report Window
The menu bar a the top of the Report Window (see Figure 1) contains
four menus. There are also some command buttons at the bottom of the
window for convenience that are also menu commands.
Menu notation
In the following menus, selections that are sub-menus are
indicated by a '![]() '. Selections prefaced with a '
'. Selections prefaced with a '![]() ' and indicate '
' and indicate '![]() ' indicate that the command is a checkbox
that is enabled and disabled respectively. Selections prefaced with a
'
' indicate that the command is a checkbox
that is enabled and disabled respectively. Selections prefaced with a
'![]() ' and indicate '
' and indicate '![]() ' indicate that the command is a
multiple choice "radio button" that is enabled and disabled
respectively, and that only one member of the group is allowed to be
on at a time. The default values set for an initial database are shown
in the menus. Selections that are not currently available will be
grayed out in the menus of the running program. The command short-cut
notation C-key means to hold the Control key and then
press the specified key.
' indicate that the command is a
multiple choice "radio button" that is enabled and disabled
respectively, and that only one member of the group is allowed to be
on at a time. The default values set for an initial database are shown
in the menus. Selections that are not currently available will be
grayed out in the menus of the running program. The command short-cut
notation C-key means to hold the Control key and then
press the specified key.
4.1 File menu
These commands are used to open the samples to be paired and other
file operations. The current menus and the menu commands (non-working
commands have a '*' prefix) are listed below. You can use either the
"Edit options" button to popup the Options Window editor to change the
input samples or the (File menu | Open Rsample) and (File
menu | Open Sample) commands.
------------------------------------
------------------------------------
------------------------------------
------------------------------------
------------------------------------
------------------------------------
![]() - update Landmark
programs and data from open2dprot.sourceforge.net/Landmark
server
- update Landmark
programs and data from open2dprot.sourceforge.net/Landmark
server
------------------------------------
4.2 Edit menu
These commands are used to change various defaults. These are saved
when you save the state and when you exit the program.
![]() Don't edit Rsample landmarks - prevent the user from changing
the set of coordinates for the Rsample landmarks. Normally, you
would not edit the Rsample landmarks after their initial definition.
This lets you use exactly the same landmarks when landmarking
other samples in subsequent sessions.
Don't edit Rsample landmarks - prevent the user from changing
the set of coordinates for the Rsample landmarks. Normally, you
would not edit the Rsample landmarks after their initial definition.
This lets you use exactly the same landmarks when landmarking
other samples in subsequent sessions.
![]() Use Rsample landmarks as fixed template - when landmarking
a new sample, the landmarks from the template Rsampe (see above)
will be available for you to match in the Sample (right image).
This ensures that all samples will be paired with the same
landmarks in the Rsample.
Use Rsample landmarks as fixed template - when landmarking
a new sample, the landmarks from the template Rsampe (see above)
will be available for you to match in the Sample (right image).
This ensures that all samples will be paired with the same
landmarks in the Rsample.
------------------------------------
------------------------------------
![]() - change the command
line options
- change the command
line options
4.3 View menu
This menu contains commands to inspect Landmark database data.
------------------------------------
------------------------------------
4.4 Help menu
These commands are used to invoke popup Web browser documentation on
Landmark. Some of the commands will load local documentation in the
the GUI report window.
![]() - this
reference manual
- this
reference manual
--------------------------------------
--------------------------------------
5. The popup Landmark Editor Window

Figure 3. Screen view of the popup Landmark Editor Window.
This lets you create or edit landmarks defined to
be common spot positions between the two samples. One of the samples
is the Rsample (on the left) and the sample to be landmarked with it
on the right (Sample). You select the (Rsample, Sample) in the main
Landmark Report Window (see Figure 1). The editor window shows four
images. The unmagnified view of Rsample and Sample are on the lower
row of images and the magnified corresponding images are on the top
row. There is a text area at the top for messages to the user. There
is a set of pull-down menus at the top File, View,
Edit, and Help. A row of controls on the bottom helps
speed up landmarking. There is a 1X to 6X zoom on the left that only
zooms the upper images. After you have clicked on corresponding spots
in the Rsample and Sample (lower images), select the Flicker checkbox
(with (C-F) short-cut) to alternately display the upper Rsample
and Sample images in the upper right image. You can use this to verify
the spots are probably the same between the two samples. The flicker
rate is determined by the delay scrollers at the bottom. The Add
LM button (with (C-A) short-cut) will add these two
selected coordinates as a new landmark spot. Similarly, by first
selecting a landmark spot with the mouse and then pressing the
Delete LM button (with (C-D) short-cut) it will delete
this landmark from the list. See the detailed menu commands below for
other information on landmarking usage.
5.1 Editor File menu
![]() Log text output to Report window - copy text output to
Report window.
Log text output to Report window - copy text output to
Report window.
------------------------------------
5.2 Editor View menu
![]() Flicker Rsample vs Sample
(C-F) - flicker the Rsample and Sample zoom windows (top
row) into the upper right zoom window. The flicker rate is
determined by the delay scrollers at the bottom.
Flicker Rsample vs Sample
(C-F) - flicker the Rsample and Sample zoom windows (top
row) into the upper right zoom window. The flicker rate is
determined by the delay scrollers at the bottom.
![]() Add landmarks radii overlay - add half-radii overlays
for the defined landmarks
Add landmarks radii overlay - add half-radii overlays
for the defined landmarks
![]() Show radii table data for landmark spot - show additional
numeric data for a landmark spot in the text area when you click
on a landmark.
Show radii table data for landmark spot - show additional
numeric data for a landmark spot in the text area when you click
on a landmark.
![]() Use +# instead of +
Use +# instead of +
------------------------------------
![]() Use mouse draw to select spots - instead of having to click
on a spot
Use mouse draw to select spots - instead of having to click
on a spot
![]() Add focus box to aid
landmarking - draw a "focus" box around the area you are
editing in the Rsample.
Add focus box to aid
landmarking - draw a "focus" box around the area you are
editing in the Rsample.
------------------------------------
![]() Separate Flicker Window
- use a separate flicker window instead of the upper left image
windows. You may resize this separate window to see more
detail. It is more useful on higher resolution monitors.
Separate Flicker Window
- use a separate flicker window instead of the upper left image
windows. You may resize this separate window to see more
detail. It is more useful on higher resolution monitors.
------------------------------------
![]() - select
the guard region color. This addes a colored region (1/4 of the
height or width) to the the image on all four edges so that you
can scroll and center the image to data on any of the four
edges.
- select
the guard region color. This addes a colored region (1/4 of the
height or width) to the the image on all four edges so that you
can scroll and center the image to data on any of the four
edges.
5.3 Editor Edit menu
![]() Don't edit Rsample landmarks).
Don't edit Rsample landmarks).
5.4 Editor Help menu
6. Downloading, installing and running Landmark
The installation packages are available for
download from the SourceForge Files mirror. Look for the most
recent release named "Landmark-dist-V.XX.XX.zip". These releases include
the program (both as Windows .exe file and a .jar file), required jar
libraries, demo data, Windows batch and Unix
shell scripts. Download the zip file and put the contents where you want
to install the program. Note that there is a Landmark.exe
(for Windows program). You might make a short-cut to this to use in more easily
starting the program. Alternatively, you can use the sample .bat and .sh scripts
to run the program explicitly via the java interpreter. Note that this method
assumes that you have Java installed on your computer and that it is at
least JDK (Java Development Kit) or JRE (Java Runtime Environment)
version 1.5.0. If you don't have this, you can download the latest version free
from the java.sun.com Website.
![]()
Figure 6. Startup icon for Landmark.
Clicking on the icon starts Landmark. To start Landmark, click on the startup
icon shown in Figure 6 below - or you can run the demo-Landmark.bat
script. For Unix systems including MacOS-X, you can start Landmark from the
command line by clicking on the Landmark.jar file or using the
demo-Landmark.sh script. These two scripts run the program in batch
but use a GUI. You could make short-cuts (Windows)
or symbolic-links in Unix to make it easier to start.
6.1 Requirements: minimum hardware and software requirements
A Windows PC, MacIntosh with MacOS-X, a Linux computer or a Sun
Solaris computer having a display resolution of at least
1024x768. We find that a 1024x768 is adequate, but a 1280x1024 screen
size much better since you can also see the Popup Report window,
Options window, and Landmark, ROI or Calibration window at the same
time. At least 30 Mb of memory available for the application is
required and more is desirable for comparing large images or
performing transforms. If there is not enough memory, it will be
unable to load the images, the transforms may crash the program or
other problems may occur.6.2 Files included in the download
The following files are packaged in the distribution you install.
you can periodically a (File | Update from Web server | ...
program) menu command to update the files from the open2dprot.sourceforge.net
Web server.
You can do a (File | Update from Web server | Landmark demo files)
menu command to update it.
7. List of the command line switches
The command line usage is:
Landmark -rsample:reference-sample -sample:sample [< optional switches >]
where the order of arguments is not relevant. In the following list,
items in bold are specific values which must be used (e.g., for
-inputFormat:{X | T}, whereas variable values in
italics indicate that a numeric value for that variable should
be used. Some switches have several alternate fixed choices in which
case this indicated as a list of bolded items inside of a set of
'{...}' with '|' separating the items. You must pick one of the items
and do not include the '{}' brackets. Also, do NOT include any extra
spaces in the arguments of the switch - it will be counted as if it
were another switch.
Command line switches
-accessionFile:accFile to overide the default accession
database file. (Default is -noaccessionFile:accession.xml).
-backupDatabases makes backups of the Landmark and landmark databases "+
"if they are edited. (Default is -commutativeLMS).
-commutativeLMS lets you search the landmark database for sample pairs
where the Rsample and Sample are swapped. (Default is
-commutativeLMS).
-debugBits:bits,optLandmarkNbr dumps various conditional
debugging parameters onto the report window. The 'bits' are the
debug bits specified as either octal or decimal and enable
particular debugging output if the program was compiled with
debugging enabled. (Default is -nodebug).
-default sets the default switches to a specific configuration:+
-nodemo
-inputFormat:X -projDir:demo/,
-LandmarkDB:Landmark.xml.
This disables -demo if it was set. (Default -nodefault).
-demo sets the default switches and sample input sample to a specific
configuration. This may be overriden by turning off the -demo
switch in the Options Wizard.
-dtd adds the XML DTD file (Open2Dprot-SPF.dtd) in the output XML
if -spfFormat:X is set. (Default is -nodtd).
-gui to start the spot pairing with a popup Graphical User Interface
rather than in batch mode. Landmark does not run in batch
mode. This captures messages from Landmark. You can then cut
and paste the results or save it to a text file. The GUI is
also used to change the switch options, re-run the spot pairing
and view images after each analysis. (Default is -gui).
-inputFormat:
7.1 Examples of some typical sets of switches
The following shows a few examples of useful combinations of command
line switches.
Any case-independent switch may be negated by preceeding it with
a 'no' eg. '-notimer'.
The command line syntax used to invoke it is:
Landmark [< opt.-switches >]
The following examples using switches might be useful:
Landmark -gui -project:demo/
# Start Landmark and load the Landmark database from the demo project.
Landmark -rsample:gel-HM-019 -sample:gel-HM-071 -gui -project:demo/
# Start Landmark with two samples already in the Landmark database
8. Demonstrations
8.1 Examples - samples of screen shots
To give the flavor of running the spot pairing program, we provide a
few screen shots of the graphical user interfaces and some images
generated by the program. 8.2 Example - output of the Report Window for ...
The following Report Window output was generate for the images in the
above example.
8.3 Examples - using command line processing for batch
It is possible to run the Landmark from the command line in your
operating system. We give two examples doing this. The first example
shows a script for the Microsoft Windows batch (.bat) file
for processing 3 images with the same reference image demo-Landmark.bat file (available on the Files
Mirror. The second example
shows the same commands in a shell script for a Unix operating system
(Linux, MacOS, Solaris, etc.) is available at from the Files mirror at demo-Landmark.sh. 8.3.1 Examples - batch processing under Microsoft Windows
REM File: demo-Landmark.bat - interactively define landmark spots for each of 3 samples
REM when paired with the reference sample.
REM This example assumes that all .jar files listed below and demo/ directory are
REM in the current directory. Modify for other situations.
REM
REM The JDK should be installed and version 1.4 or later is required.
REM You can download the latest JDK from http://java.sun.com/
REM
REM The files needed are listed below:
REM JAR files required and mentioned in manifest:
REM xml-apis.jar xercesImpl.jar jai_codec.jar jai_core.jar O2Plib.jar
REM
REM demo image files:
REM demo/ppx/gel-HM-019 (Reference sample)
REM demo/ppx/gel-HM-071
REM demo/ppx/gel-HM-087
REM demo/ppx/gel-HM-096
REM Accession database file is in:
REM demo/xml/accession.xml
REM Landmark database file is in:
REM demo/xml/landmark.xml
REM Sample Spot-list files (SSF) are in:
REM demo/xml/gel-HM-019-SSF.xml
REM demo/xml/gel-HM-071-SSF.xml
REM demo/xml/gel-HM-087-SSF.xml
REM demo/xml/gel-HM-096-SSF.xml
REM Generated images are saved in:
REM demo/tmp/
REM
REM P. Lemkin $Date$
echo "demo-Landmark.bat"
pwd
date /T
java -Xmx256M -jar Landmark.jar -demo -project:demo\ -sample:gel-HM-071 -rsample:gel-HM-019
java -Xmx256M -jar Landmark.jar -demo -project:demo\ -sample:gel-HM-087 -rsample:gel-HM-019
java -Xmx256M -jar Landmark.jar -demo -project:demo\ -sample:gel-HM-096 -rsample:gel-HM-019
echo "-- Finished landmarking the samples ---"
date /T
8.3.2 Examples - batch processing under Unix (or MacOS-X)
Because java is relatively operating system independent, the same java
command lines are used with the "\" changed to "/", "REM" changed to
"#", and "DATE/T" to "date" from Windows to
Unix script and file path conventions.
#!/bin/sh
# File: demo-Landmark.sh - interactively define landmark spots for each of 3
# samples when paired with the reference sample.
# This example assumes that all .jar files listed below and demo/ directory are
# in the current directory. Modify for other situations.
#
# JAR files required and mentioned in manifest:
# xml-apis.jar xercesImpl.jar jai_codec.jar jai_core.jar O2Plib.jar
#
# The files needed are listed below:
# JAR files required and mentioned in manifest:
# xml-apis.jar xercesImpl.jar jai_codec.jar jai_core.jar O2Plib.jar
#
# demo image files:
# demo/ppx/gel-HM-019 (Reference sample)
# demo/ppx/gel-HM-071
# demo/ppx/gel-HM-087
# demo/ppx/gel-HM-096
# Accession database file is in:
# demo/xml/accession.xml
# Landmark database file is in:
# demo/xml/landmark.xml
# Sample Spot-list files (SSF) are in:
# demo/xml/gel-HM-019-SSF.xml
# demo/xml/gel-HM-071-SSF.xml
# demo/xml/gel-HM-087-SSF.xml
# demo/xml/gel-HM-096-SSF.xml
# Generated images are saved in:
# demo/tmp/
#
# P. Lemkin $Date$
echo "demo-Landmark.sh"
pwd
date
java -Xmx256M -jar Landmark.jar -demo -project:demo/ -sample:gel-HM-071 -rsample:gel-HM-019
java -Xmx256M -jar Landmark.jar -demo -project:demo/ -sample:gel-HM-087 -rsample:gel-HM-019
java -Xmx256M -jar Landmark.jar -demo -project:demo/ -sample:gel-HM-096 -rsample:gel-HM-019
echo "-- Finished landmarking the samples ---"
date
9. Landmark References
These papers (a subset of the GELLAB-II papers),
reference the GELLAB-II spot pairing program. The Open2Dprot
Java-language Landmark program was other Open2Dprot programs and
concepts were used from the old GELLAB-II C-language program as well
as from code from the MAExplorer and Flicker projects.
This program will be replaced with a full XML editor that will make
use of the new MIAPE standard. New Java code was added as
well. Although Landmark has been enhanced in many ways, the basic
algorithm is similar so these papers may be useful for more details on
the algorithm.
| Contact us | Landmark is a contributed program available at
open2dprot.sourceforge.net/Landmark
Powered by |
Revised: 05/19/2006 |