Notes on the current Open2Dprot XML
data schemas
(*** PRELIMINARY AND
NOT COMPLETE - WORKING DOCUMENT ***)
Created: 3-27-2005
Revised: 3-28-2005
Revised: 12-17-2005
Peter F. Lemkin
CCRNP, CCR, NCI
Frederick, MD 21702 USA
E-mail: lemkin@users.sourceforge.net
http://open2dprot.sourceforge.net/
Table of contents
1. Introduction to use of XML in Open2Dprot
2. computing
environment
3. Open2Dprot pipeline
processing modules and scheduler
3.1 Classes of data
objects used in Open2Dprot
4. The use of standard
XML interchange
5. The Open2Dprot
project list of XML schemas
5.1
Sample accession XML data schema
5.1.1 Fields in
Accession XSD (Open2Dprot-Accession.xsd)
5.1.2 Required
computable data fields for each <SampleEntry>
5.1.3 Experiment
information data fields for each <SampleEntry>
5.2 Landmark spot data XML schema between two
samples
5.2.1 Fields in
Landmark XSD (Open2Dprot-Landmark.xsd)
5.2.2 Definition of a
<LandmarkSet> entry
5.3
Sample spot-list data (SSF) XML schema
5.3.1 Fields in sample
spot-list XSD (Open2Dprot-SSF.xsd)
5.3.2 SSF Preface
<Sample_parameters> schema
5.3.3 SSF spot data
<Spot> schema
5.3.4 SSF Epilogue
statistics <Global_segmenter_statistics> schema
5.4
Paired sample spot-list data (SPF) XML schema
5.4.1 Fields in sample
paired-spot-list XSD (Open2Dprot-SPF.xsd)
5.4.2.1 Contents of
the <Rsample> and <Sample> objects in CSD schema
5.4.2 SPF Preface
<Pairing_parameters> schema
5.4.3 SPF paired-spot
<Pspot> schema
5.4.4 SPF Epilogue
<Global_Spot_pairing_statistics> schema
5.5 Composite Sample Database (CSD) XML
schema (Open2Dprot-CSD.xsd)
5.5.1 The canonical
sample, Csample, for replicate samples in the CSD
5.5.2 The expression
data for a single spot in the CSD or Rspot
5.5.3 A
post-translational modification Rspot or <PTMRspot>
5.5.4 The
<ConditionsList> as a list of <Conditions> groups of samples
1. Introduction to use of XML in Open2Dprot
The following notes describe the way we are currently using XML
data for interchange in Open2Dprot (http://open2dprot.sourceforge.net/).
This document also discusses how we would like to take advantage of the
Proteomics Standards Initiative (PSI at http://psidev.sourceforge.net/)
MIAPE, GelML and related parts of the General Proteomics Standards (GPS) when they
stabilize.
We are migrating our XML coding from using the Apache Xerces http://xerces.apache.org/ SAX XML reader
and hand-coded XML writers to using XMLbeans (http://xmlbeans.apache.org/). This
document describes the current implementation
The Open2Dprot project is a community effort to create an open-source n-dimensional (n-D) protein expression data analysis system. It will be downloadable and could be used for data mining protein expression across sets of n-D data from research experiments. Modules will be created for
2-dimensional data including 2D-PAGE (polyacrylamide gel electrophoresis) and initial support for 2D LC-MS, protein arrays and other data separation methods.
Our goal, in using an XML data interchange format, is to be as
compliant as possible with MIAPE. For those cases where we have parameters and
summary statistics that are not currently MIAPE compliant and there is an
escape mechanism in MIAPE to encode this information, we will refactor our
current XML to use that mechanism. Otherwise, we will use additional schemas to
fill the gaps.
At the Montreal 2003 HUPO meeting, Chris Taylor (EBI) suggested we
go ahead and implement our own schema while MIAPE was being developed. We used
what was available and we needed that already existed in the PEDRo schema and
then added fields we needed that were missing from PEDRo.
In addition, when a more complete MIAPE model is available that
handles these additional complex types; we will then refactor our remaining
code and new data-mining code and schemas toward that model.
Note that the current Open2Dprot XML schemas we are presenting
here are placeholders to be redone when the new GelML/MIAPE standard is
available and meets our needs. At that time we plan to go back and refactor the
code to take the new standard into account.
The files discussed throughout this document are available on the http://open2dprot.sourceforge.net/
server. We will specify the direct links to specific files
throughout the document to make it easier to review the files. Documentation
(manuals, PDFs, javadocs), source code (CVS and Files mirror releases), demo
data, and downloadable installers are available on the Web site.
The primary concept of pipeline processing for the Open2Dprot
system is to construct a Composite Samples Database (CSD) of protein expression
values of corresponding proteins across multiple samples. Once this CSD
database is constructed, it can then be used for subsequent analysis.
Section 2 describes
the Open2Dprot computing environment shown in Figures 1,
2, and through 3
that follow. Section 3 describes the
pipeline-processing paradigm. Section 4
discusses our goal of using common XML standards to facilitate data
interchange. Section 5 describes
our current XML schemas.
Note that this is a work-in-progress. We are in the process of
defining the XML schema for the CSD and so dont have the full XML schema we
will use. However, we outline some of the key export data of an assembled CSD
as follows in Section 5.5.
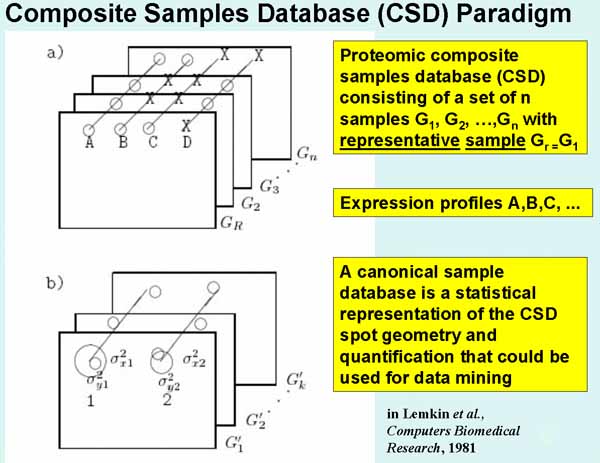
Figure 1. Composite Samples Database model, CSD, used in Open2Dprot.
a) Illustrates a composite sample database. Corresponding
paired spots (circles) are denoted by diagonal lines drawn through them. Such
sets of corresponding spots are called Rspot sets. One of the samples (from the
set of samples {GR, G2, G3, ..., Gn}
is selected to be a reference sample or Rsample, denoted GR. The
circle means the spot is present and the X means that it is missing in that
sample. Spot A occurs in all n samples. Spot B occurs in the Rsample and in one
other sample. Spot C is only in the Rsample. Spot D is not present in the
Rsample but is in most of the other samples. Spots A, B, and C are in the
un-extended Rspot database (since they occur in the Rsample) while spot D is in
the eRspot part of the database (since it does not occur in the Rsample). Part
of constructing the CSD is to extrapolate where missing spots would be in the samples
that they are missing based on their relationship to neighboring spots common
to the all samples. Extrapolated spots are assigned expression and area values
of 0 and can be used for "missing spots" types of tests. Although the
CSD in the initial Open2Dprot project, is constructed using a common reference
sample, it is be possible to construct the CSD using a transitive model mapping
across pairs of samples - although with higher error rates. b) Illustrates the basis of using
mean (or median) spot positions for estimating canonical spots for a subset of
k samples from the n samples database. The canonical spot can then be used to
estimate the position of spots missing from some of the other samples. The mean
and variances of Rspot positions across a set of samples is mapped to the
coordinate system of the Rsample. When that has been done, the set of samples
of the same experimental class can be replaced by a single averaged sample
called the Csample' (the estimate of the canonical sample for a set of replicate samples). The mean displacement vector of a canonical spot from its associated
landmark spot (in any sample under discussion) is used to extrapolate the
position in samples where the expected canonical spot is missing. If no
landmark spots were used in constructing the CSD, nearby Rspots that have spots
from all samples may be used to supply the vector offsets. Similarly, a mean
quantified spot data can be used to represent a set of replicate samples.
_________________________________________________________________________________
The following figure 2 illustrates
the context of the schemas in
Open2Dprot processing. Figure 2.1 shows more of the details of the
XML schemas used between pipeline stages.
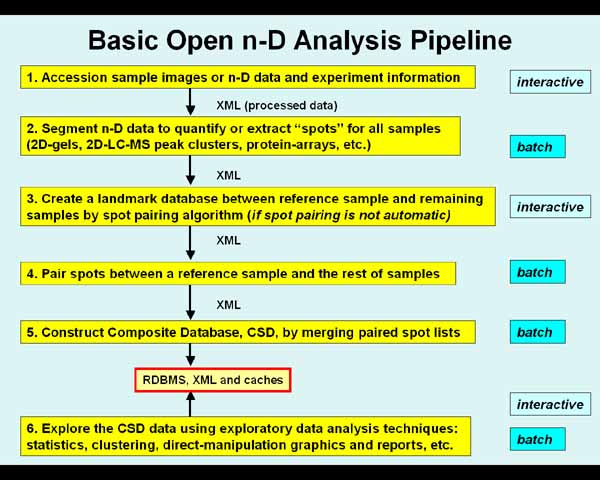
Figure 2. The Open2Dprot pipeline
processing data reduction hierarchy.
This shows the data reduction steps in converting n-D
sample image (whether real images or "virtual" images) data to a
composite sample database suitable for exploratory data analysis. In general,
each step of the pipeline depends on the previous step being completed. Some
steps can be omitted (e.g., if protein array pre-quantified spot data were used
then Steps 2-4 can be omitted since no spot segmentation is required, no
landmarking and no spot pairing is required; if a spot-pairing method were used
that did not depend on predefined landmarks, then Step 3 can be omitted, etc).
A key design element of Open2Dprot is that pipeline steps are assigned one of
several alternated modules that adhere to the same XML input/output schema.
That means that alternate methods can be substituted in the pipeline. For
example, for 2D gels an image segmenter would be used for step 2; for 2D LC-MS peak
cluster data a clustering method might be used; for a protein array spot list
data might be used from one of the many microarray image spot segmentation
programs, etc. The pipeline is set up and controlled by the Pipeline Control
Program.
______________________________________________________________________________
|
| (2D images, 2D LC-MS spot cluster data, protein array data)
v
1.
Accession sample images or n-D data and experiment information,
|
| XML (sample Accession data Open2Dprot-Accession.xsd)
v
2.
Segment n-D data to quantify or extract "spots" for all
samples (2D-gels, 2D-LC-MS, etc.),
|
| XML (sample spot-list SSF data Open2Dprot-SSF.xsd)
v
3.
Create a landmark database between reference sample and remaining
samples by spot pairing algorithm (if required for spot pairing),
|
| XML (Landmark data Open2Dprot-Landmark.xsd)
v
4.
Pair spots between a reference sample and the rest of samples,
|
| XML (samples paired-spot-list SPF data Open2Dprot-SPF.xsd)
v
5.
Construct Composite Samples Database, CSD, by
merging paired spot lists,
|
|
v
RDBMS and caches (CSD data Open2Dprot-CSD.xsd)
^
|
|
6.
Explore the CSD data using exploratory data analysis and data mining
techniques: statistics, clustering, classification, direct-manipulation
graphics, reports, etc. This may invoke Java plugins and R-language
scripts.
______________________________________________________________________________
Figure 2.1 The Open2Dprot pipeline
processing data reduction hierarchy. The pipeline processing hierarchy is
illustrated by figure 2 of the Open2Dprot home page. See the figure legend and
discussion of which stages could be run in background batch.
2. Open2Dprot computing environment
Open2Dprot is meant to be run locally on an investigator's or a
collaborative group's computer. However, the CSD database will reside on a
RDBMS that could be split between a working experiment database and a reference
database (e.g., plasma proteins, etc.) with spot identifications and other
proteomic information.
Currently, Open2Dprot saves all data (XML data interchange files
as well as derived images) from a project in a set of sub-directories in a
project directory. Multiple experiments, each consisting of multiple samples,
could reside in the same project directory. Multiple CSDs could be constructed
in the same project directory created from samples from different experiments
or subsets of samples.
<project-directory>/batch/
- batch files
<project-directory>/cache/
- cache files
<project-directory>/ppx/ - original input image files
<project-directory>/rdbms/
- RDBMS CSD database files
<project-directory>/tmp/ - generated temporary and derived image
files
<project-directory>/xml/ - accession DB, landmark DB, SSF files,
SPF files, etc.
These data could reside in the RDBMS itself or in sets of
distributed RDBMS. The cache directory data files are used for data mining
rather than paging data from the RDBMS, which would be too slow for many
exploratory data analysis operations.
3. Open2Dprot pipeline
processing modules and scheduler
There is a top-level scheduler program (not released yet) called
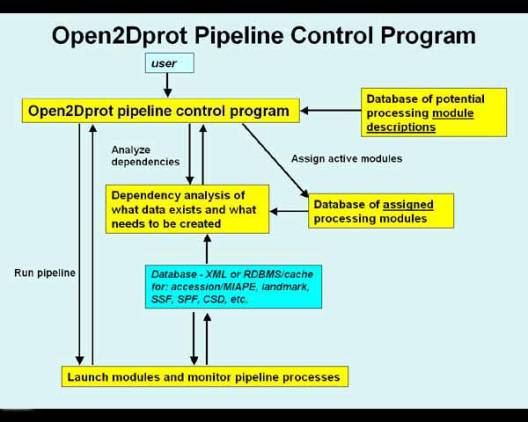
Open2Dprot illustrated in Figure 3 that determines
the data dependency, what data exists, what data is required (i.e., the next
step of the processing depends on it). It then runs the appropriate pipeline
modules to create that data.
Each pipeline step may have a specific module dynamically
assigned. This means that different pipeline processing method sets can be
created for different types of proteomic expression data. For example, if no
spot pairing is required (e.g., the input is a protein array and not a 2D gel
image or 2D LC-MS data file), then the pipeline analyzer will skip the spot
pairing steps. Similar dependencies can be assigned to all processing steps.
______________________________________________________________________________
Figure 3. The Open2Dprot pipeline
control program.
The pipeline control program (called Open2Dprot
- and is under development) will schedule and run the modules in the pipeline
after doing a data-dependency analysis on 1) what data exists, and 2) what data
needs to be created by running parts of the pipeline to proceed to the next
stage in the pipeline. That is, it works backwards from the future CSD to determine
what data is required and then repeats that analysis further back in the
pipeline until it reaches the top of the pipeline (Figure 2) or reaches data
that already exists in the pipeline. It then executes the pipeline processes to
construct the CSD. Once the CSD is created, the pipeline is no longer needed
except to add additional samples. Instead, the CSDminer program would then be
used directly to access the already existing CSD database.
All currently released pipeline modules, source code, and the
common library O2Plib are available from the Web site list of subprojects
http://open2dprot.sourceforge.net/doc/subprojects.html
The direct links are:
http://open2dprot.sourceforge.net/Accession
http://open2dprot.sourceforge.net/CmpSpots
http://open2dprot.sourceforge.net/Seg2Dgel
http://open2dprot.sourceforge.net/Landmark
http://open2dprot.sourceforge.net/O2Plib
where:
Accession - sample experiment and ROI accession
program
Seg2Dgel - segment 2D gel (or similar) image into a
Sample
Spot-list File (SSF)
Landmark - interactively define a set of landmarks
between
two sample
images (Landmark)
CmpSpots - pair two sample spot lists into a
Sample
Paired-spot-list File (SPF)
O2Plib - common Open2Dprot library
O2Plib.db.*
contains data objects and XML I/O
O2Plib.db.CSD contains CSD data
objects and XML I/O
[Modules not yet
released: Open2Dprot, BuildCSD, CSDminer
Open2Dprot Open2Dprot
pipeline scheduler
BuildCSD -
construct/add-to the CSD from sets of SPF data files
CSDminer - exploratory analysis for CSD
Although we refer to a XML Sample Spot-list File (SSF) as a file,
it could be a XML object in a RDBMS.
3.1 Primary data objects used in Open2Dprot
The primary
classes which define data objects used throughout Open2Dprot are defined in the
O2Plib.db.* Java library modules and define the base objects and their XML
readers and writers.
DbAccession.java - read, write and access
accession sample database
DbBaseSpot.java base spot class
DbBoundary.java - spot boundary manipulation
(-- not released)
DbLM.java - read, write and access paired
sample landmark spots database
DbPairSamples.java - read, write and access
paired sample instance
DbSample.java - read, write and access
sample spot list instance
DbPspot.java SPF paired-spot feature
object
DbSpot.java SSF spot feature object
LMset.java landmark set object.
The O2Plib home
page is http://open2dprot.sourceforge.net/O2Plib
It has javadoc API documentation accessible from the home page showing the object dependencies.
3.1.1 Primary data objects used in CSD for Open2Dprot
The primary
classes which define CSD data objects used throughout Open2Dprot are defined in
the O2Plib.db.CSD.* Java library modules and define the CSD base objects and
their XML readers and writers. See Section 5.5 for discussion on how these
describe the CSD XML schema.
CSD.java instance of CSD database
CSDacc.java Accession data instance
CSDannotation.java annotation instance
CSDcache.java cache instance
CSDcal.java grayscale to measurement units
calibration instance
CSDcond.java sample conditions instance
CSDexpr.java expression profile instance
CSDexprList.java list of expression
profiles instance
CSDfilterState.java data filtering
instance
CSDglb.java global state instance
CSDio.java I/O instance
CSDlimits.java filter limits instance
CSDlms.java landmark data instance
CSDnorm.java sample spots normalization
instance
CSDRmap.java reference map instance
CSDRspot.java Rspot (reference spot) set
instance
CSDRspotList.java list of Rspots instance
CSDsizes.java current size limits instance
CSDtotals.java current samples, spots etc
totals instance
4. The use of standard XML interchange data files
Open2Dprot standardizes data interchange through XML files. The
I/O is to/from data files, which in the future will also be able to be kept in
a relational database.
To minimize problems porting data between processing modules, we
have created a common Java library O2Plib. This library defines all data
structures that would be used in more than one Open2Dprot pipeline module.
It also centralizes the XML I/O.
The early, current, version of the Open2Dprot XML library uses the
SAX XML reader to read XML data files. It also uses Java methods to explicitly
generate the XML output. These used optional document type definitions (DTDs)
with the SAX readers (if the dtd command-line switch was specified with
the module).
Future XSD XMLbeans I/O
We are in the process of refactoring the XML I/O using XMLbeans (http://xmlbeans.apache.org/) generated
XML readers and writers based on Java code generating from xsd schemas. This
will make it much easier to integrate the MIAPE xsd schemas with Open2Dprot. We
will be adding xsd namespaces to the Open2Dprot schemas to keep the fields
unique. We will use the merge with MIAPE schema fields if possible. The name
space will be the following:
o2p: Open2Dprot name space that will be used if
there is no
equivalent MIAPE
schema type or element
If there is a conflict between the Open2Dprot sub-schemas and
there is no MIAPE replacement, then we may rename our elements and type names
or we might use alternate namespaces as follows. This is to be avoided if
possible by the renaming of fields.
o2pa: Open2Dprot accession space
o2pl: Open2Dprot landmark space
o2ps: Open2Dprot spot list space
o2pp: Open2Dprot paired-spots list space
o2pc: Open2Dprot CSD space
Current SAX XML readers
There are document type definitions (DTDs) associated with the XML
data readers and writers in the Library. All XML I/O is handled completely by
this library. Currently, we are using hardwired Xerces SAX readers, but hope to migrate
this to dynamic XML schema I/O this year.
Currently, we have 4 published DTDs on the Web site (the CSD will
be an XML schema file Open2Dprot-CSD.xsd and is being constructed for the
Composite Sample Database).
The DTDs are available several ways: 1) download and install the
CmpSpots program (this includes the four DTDs), or 2) they are available at:
http://open2dprot.sourceforge.net/O2Plib/Open2Dprot-Accession.dtd
http://open2dprot.sourceforge.net/O2Plib/Open2Dprot-Landmark.dtd
http://open2dprot.sourceforge.net/O2Plib/Open2Dprot-SSF.dtd
http://open2dprot.sourceforge.net/O2Plib/Open2Dprot-SPF.dtd
5. The Open2Dprot project list of XML schemas
Once the Composite Sample Database (CSD) is constructed it could
be used for exploratory data analysis and data mining. It consists of a MIAPE
sample experiment data and a list of paired-"spot" expression values
across all samples in the database. Missing sample spot values are allowed and
are described in Section 5.5.
The CSD database is viewed as a multiple-sample database is
illustrated in Figure_1_CSD in the Introduction to
this document and discussed in the home page of the Open2Dprot Web site.
There are other types of informatics data that are both useful and
necessary such as sets (i.e. subsets) of spots, condition sets (subsets of
samples), calibration data for each sample (as different from normalization),
etc.
The complex XML types definitions are highlighted in yellow in
this Section. E.g., see the definition of <CSD> in Section 5.5.
5.1 Sample accession data XML
schema
A single sample is a 2D gel image, a 2D LC-MS data set, a protein
array, or some other quantitative or semi-quantitative protein expression
vector etc. A DIGE type sample is really a set of samples so each channel is a
separate sample - even though spots are effectively paired by the way the
samples are run simultaneously in the same gel with different dyes.
Each sample has some input data (e.g., 2D gel image, 2D LC-MS real
image, 2D LC-MS clustered peaks data with a virtual image, protein array image
or protein array spot list in some format), etc. The sample name is currently
the name of the data file without a path and without the extension (e.g., .tif,
.jpg, .gif, etc). The extension is
determined at run time.
The accession database contains enough information to find the
sample, grayscale or other calibration information, regions of interest, etc.
The Accession program currently creates and edits this
information. We plan to replace this program with a more advanced program to
fill in the other MIAPE experiment information fields.
The accession database DTD is available at
http://open2dprot.sourceforge.net/O2Plib/Open2Dprot-Accession.dtd
The accession database XSD is available from the Open2Dprot Web
site CVS server at
http://cvs.sourceforge.net/viewcvs.py/open2dprot/schemas/Open2Dprot-Accession.xsd?rev=1.14&view=log
There is an example of an accession XML file in the file
http://open2dprot.sourceforge.net/demo/xml/accession.xml
5.1.1 Fields in the Accession XSD (Open2Dprot-Accession.xsd)
Samples are accessioned (i.e., entered) into a database containing
experiment information about a sample as well as global image descriptions
(file name, size, data region-of-interest (ROI), pixel grayscale to measurement
units calibration, etc). In the case of non-image samples (e.g., 2D LC-MS peaks
data, protein array data), these fields are defined for the virtual image of
that data.
Additional experiment related information is also saved in the
accession database for each sample. This includes study, investigator, date,
sample conditions, etc.
This is currently defined using the Open2Dprot common library
class O2Plib.db.DbAccession.java.
The accession database file
<project-directory>/xml/accession.xml
contains sample and experiment information in a single flat-file.
It is similar to a subset of the original PEDRo
proteomics schema. [We note that that this list of sample description
accession data descriptors is inadequate for many research purposes and
will be replaced with a more general MIAPE set of descriptors.] We
currently use this accession schema as a placeholder until we implement a full
MIAPE sample experiment information subset.
We would of course have to integrate the GUI for the ROIs (Region
Of Interest) and grayscale calibrations.
A <SampleEntry> consists of required computable data
fields, and
experiment information data fields in the subsections that follow.
The Accession
pipeline module program allows the creation and editing of the accession sample
database. We plan on making the Accession module more MIAPE compliant by either
extending Accession or replacing it with another accessioning program. The home
page is
http://open2dprot.sourceforge.net/Accession
The accession database DTD is available at
http://open2dprot.sourceforge.net/O2Plib/Open2Dprot-Accession.dtd
The accession database XSD is available from the Open2Dprot Web
site CVS server at
http://cvs.sourceforge.net/viewcvs.py/open2dprot/schemas/Open2Dprot-Accession.xsd?rev=1.14&view=log
There is an example of an accession XML file in the demonstration
directory
http://open2dprot.sourceforge.net/demo/xml/accession.xml
The top-level object contains two database identifier fields and
an arbitrary list of <SampleEntry>s.
<Accession>
<DatabaseName> (String) name of the accession database
</DatabaseName>
<Date> date database was created or modified</Date>
<SampleEntry> sample entry 1 </SampleEntry>
<SampleEntry> sample entry 2 </SampleEntry>
. . .
<SampleEntry> sample entry n </SampleEntry>
</Accession>
The <SampleEntry> complex type contains two types of data:
required computable data and optional experiment annotation data. These are
described in Sections 5.1.2 and 5.1.3.
5.1.2 Required computable data fields for each <SampleEntry>
The accession data contains several critical computable data
fields for each <SampleEntry>. These fields are required by any
Open2Dprot programs that need to lookup accession information for one or more
samples.
In particular, the spot segmentation module, Seg2Dgel, requires a
region of interest (ROI) called the computing window where spots will be found.
If this ROI is not defined, it is defined as the entire image or (x,y) space of
the data (in the case of abstract 2D LC-MS peak data or protein arrays. If a
grayscale calibration is present, it will calibrate grayscale in terms of the
calibration data. Fields that have optional entries are indicated with [opt].
<SampleEntry>
<Sample> (String) sample base name (if there is an image, no path
or
extension)
</Sample>
<Rsample> (String) reference sample base name (if ANY) (if there
is
an image, no path or extension)
</Rsample>
<WedgeCalList> (String) optional calibration values list that is
synchronized with <WedgeGrayList>. The delimiters
may be commas,
spaces, or tabs.
</WedgeCalList>
<WedgeGrayList> (String) optional corresponding grayscale peak
values list
synchronized with <WedgeCalList>. The
delimiters may be commas,
spaces, or tabs.
</WedgeGrayList>
<cwx1>(int) X ULHC of computing region of interest (ROI) where
spots
reside in the image or data object in the Cartesian
raster
system [x1:x2, y1:y2] in pixels. (0,0) is ULHC of
image.
Spots outside of ROI could be ignored if analysis
module
uses the ROI.
</cwx1>
<cwy1>(int) Y ULHC of computing region of interest (ROI)
</cwy1>
<cwx2>(int) X LRHC of computing region of interest (ROI)
</cwx2>
<cwy2>(int) Y LRHC of computing region of interest (ROI)
</cwy2>
<calCWx1> (int) X ULHC of optional calibration wedge region of
interest
ROI (e.g., optical density (OD) step wedge or some
other calibration
wedge) in pixels. A histogram of image pixels (for
images) could be
computed in this region. The peaks could then be used
to calibrate
grayscale in terms of wedge calibration units. The
wedge is defined
in the
Cartesian coordinates raster system
[calCWx1:calCWx2, calCWy1:calCWy2]. (0,0) is image
ULHC.
</calCWx1>
<calCWy1> (int) Y ULHC of optional calibration wedge ROI
<calCWy1>
<calCWx2> (int) X LRHC of optional calibration wedge ROI
<calCWx2>
<calCWy2> (int) Y LRHC of optional calibration wedge ROI
<calCWy2>
<PixWidth> (int) is image or data object width in pixels
</PixWidth>
<PixHeight> (int) is image or data object width in pixels
</PixHeight>
(.
. . Also includes optional data described in Section 5.1.3 . . .)
</SampleEntry>
5.1.3 Experiment information data fields for each
<SampleEntry>
The accession data also contains several sample
experiment-information fields for each <SampleEntry>. NOTE these
fields are probably available under other names in MIAPE and will be deprecated
when the MIAPE schema is used.
<SampleEntry>
<PatientNbr> is patient
number associated with the <Sample> </PatientNbr>
<Study> is the
experimental condition information for the <Sample> </Study>
<ExperimentDate>
(String) [opt] date the <Sample> was created or the sample
accessioned into the database.
</ExperimentDate>
<CultureReagent>
(String) [opt] culture reagents used on the <Sample>.
</CultureReagent>
<AmpholyteAndGelGradientRange> (String) [opt] ampholyte range and
gel gradient
range associated with the
<Sample>.
</AmpholyteAndGelGradientRange>
<IntervalBeforeLabeling> (String) [opt] interval before labeling
the <Sample>.
</IntervalBeforeLabeling>
<LabelingIsotope>
(String) [opt] labeling isotope associated with the <Sample>.
</LabelingIsotope>
<DurationLabel>
(String) [opt] duration of pulse-chase label associated with
the
<Sample>.
</DurationLabel>
<DurationExposure>
(String) [opt] duration of exposure (e.g., autoradiograph, etc.)
associated with
the <Sample>.
</DurationExposure>
<Camera>(String) camera, scanner or input
device used along with resolution and
0 other settings
</Camera>
<Investigator>
(String) [opt] investigaor associated with the
creation
of or who is responsible for the <Sample>.
</Investigator>
(.
. . Also includes required data described in Section 5.1.2 . . .)
</SampleEntry>
5.2
Landmark spot data XML schema between two samples
A landmark spot is a spot or (x,y) coordinate pair that is either
manually or automatically determined to be the same spot in two sample image or
virtual images 2D coordinate space.
Some spot pairing programs may require landmarks (e.g., CmpSpots) whereas other
spot pairing programs may not. In the latter case, landmark data is not
required and therefore the pipeline step to create landmarks is omitted in the
pipeline specification created or edited by the pipeline control program. The SSF data itself may implicitly not require landmarks
(e.g., if the SSF data was from a protein array or other self-identifying data
set, etc).
The landmark database DTD is available at
http://open2dprot.sourceforge.net/O2Plib/Open2Dprot-Landmark.dtd
The landmark database XSD is available from the Open2Dprot Web
site CVS server at
http://cvs.sourceforge.net/viewcvs.py/open2dprot/schemas/Open2Dprot-Landmark.xsd?rev=1.14&view=log
There is an example of a landmark XML file in the demonstration
directory
http://open2dprot.sourceforge.net/demo/xml/landmark.xml
The Landmark
pipeline module program allows the creation and editing of a set of landmarks
for pairs of samples that is stored in the landmark database.
5.2.1 Fields in the Landmark XSD (Open2Dprot-Landmark.xsd)
This is defined using the classes O2Plib.db.LMset.java,
O2Plib.db.DbLM.java, and O2Plib.db.DbSample.java.
The landmark database file
<project-directory>/xml/landmark.xml
contains pairs of sample (x,y) coordinates and landmark ids and is
a single flat-file. This is only used with software that requires
landmarks. The top-level object contains two database identifier fields and an
arbitrary list of <LandmarkSet>s. A <LandmarkSet> is one landmark
entry for an Rsample, Sample and a particular landmark. For a set of landmarks
for a given Rsample and Sample there will be a many <LandmarkSet> entries
one for each common landmark.
<LandmarkDB>
<DatabaseName> (String) name of the database </DatabaseName>
<Date> (String) date the file was created or modified</Date>
<LandmarkSet>landmark set 1 </LandmarkSet>
<LandmarkSet>landmark set 2 </LandmarkSet>
. . .
<LandmarkSet>landmark set n </LandmarkSet>
</LandmarkDB>
5.2.2 Definition of a <LandmarkSet> entry
Each pair of samples (<Sample>, <Rsample>) has a
<LandmarkSet> entry. For example, if there were 20 landmarks for the
sample pair, then there would be 20 <LandmarkSet> entries in the landmark
database for the same sample pair. The difference between the entries would be
the <lmNbr> keys which would be different (as well as the (x,y) data,
which would be different). Depending on the spot merging method used when
constructing the CSD, there may or may not be a requirement to use the same
landmarks for all samples (see Figure 2.b caption for short discussion on
this).
<LandmarkSet>
<Sample> (String) sample base name </Sample>
<Rsample> (String) sample base name of reference
sample</Rsample>
<lmNbr> (int) landmark number identifier counting from
1</lmNbr>
<xRsample> (int) X coordinate of lmNbr'th Rsample landmark
</xRsample>
<yRsample> (int) Y coordinate of lmNbr'th Rsample landmark
</yRsample>
<xSample> (int) X coordinate of lmNbr'th sample landmark
</xSample>
<ySample> (int) Y coordinate of lmNbr'th sample landmark
</ySample>
</LandmarkSet>
5.3 Sample spot-list data XML
schema (SSF)
The Sample Spot-list File (SSF) is created by processing the raw
sample data linked from the accession database. We have one example of the
pipeline module that produces this data, Seg2Dgel, which finds
spots in an image: 2D gel or 2D LC-MS (low resolution) images. The spot-list
DTD contains a list of spots and also parameters used in computing the spot
list and statistics on the spot features.
The SSF should be extended to include more-specific 2D LC-MS and
protein array fields. In the case of protein arrays, the protein
identifications should not be part of the SSF itself. Instead, a
separate identification database should contain that information. This could be
the CSD <Rmap> and <RmapList> complex types described in Section 5.5.
5.3.1 Fields in the sample spot-list XSD (Open2Dprot-SSF.xsd)
This is defined using the classes O2Plib.db.DbSpot.java and
O2Plib.db.DbSample.java.
Each sample will have a processed standardized spot list we call a
Sample Spot-list File (SSF). The SSF DTD is available at
http://open2dprot.sourceforge.net/O2Plib/Open2Dprot-SSF.dtd
The SSF database XSD is available from the Open2Dprot Web site CVS
server at
http://cvs.sourceforge.net/viewcvs.py/open2dprot/schemas/Open2Dprot-SSF.xsd?rev=1.14&view=log
The Sample Spot-List File (SSF) is saved as
<project-directory>/xml/<Sample>-SSF.xml
A SSF is described below.
Thee following are some examples of SSF XML files in the
Open2Dprot demonstration directory
http://open2dprot.sourceforge.net/demo/xml/gel-HM-019-SSF.xml
http://open2dprot.sourceforge.net/demo/xml/gel-HM-071-SSF.xml
http://open2dprot.sourceforge.net/demo/xml/gel-HM-087-SSF.xml
http://open2dprot.sourceforge.net/demo/xml/gel-HM-096-SSF.xml
The <SSF> complex type consists of these three types of
objects: preface, spot data, and epilogue.
<SSF>
<Sample_parameters> sample info and parameters
</Sample_parameters>
<Spot> spot #1 </Spot>
<Spot> spot #2 </Spot>
. . .
<Spot> spot #n </Spot>
<Global_segmenter_statistics> summary statistics
</Global_segmenter_statistics>
</SSF>
These fields are described as follows:
5.3.2 SSF Preface <Sample_parameters> schema
The <Sample_parameters> object contains sample experiment
information and parameters. Note: the full experiment information is found in
the accession database and is not repeated here.
<Sample_parameters>
<Sample_Name> (String) name of sample </Sample_Name>
<Simple_FileName> (String) input data file name. This could be an
image
file or some other data type depending on the spot
quantification program.
</Simple_FileName>
<Project_directory> (String) name of project directory where the
ppx/
input image subdirectory for the input image data
file is found.
</Project_directory>
<date> (String) date the
spot list was created </date>
<Open2Dprot_SSF_Version> (String) version of SSF schema version
</Open2Dprot_SSF_Version>
<cwx1> (int) X ULHC of computing window ROI to segment in pixels
</cwx1>
<cwy1> (int) Y ULHC of computing window ROI to segment in pixels
</cwy1>
<cwx2> (int) X LRHC of computing window ROI to segment in pixels
</cwx2>
<cwy2> (int) Y LRHC of computing window ROI to segment in pixels
</cwy2>
<Pix_Height> (int) image height or virtual image in pixels
</Pix_Height>
<Pix_Width> (int) image width or virtual image in pixels
</Pix_Width>
<t1Area_threshold> (float) [opt] lower area threshold spot sizing
limit.
Used in Seg2Dgel segmenter.
</t1Area_threshold>
<t2Area_threshold> (float) [opt] upper area threshold spot sizing
limit.
Used in Seg2Dgel segmenter.
</t2Area_threshold>
<t1Density_threshold> (float) [opt] lower integrated density
threshold
spot sizing limit. Used in Seg2Dgel segmenter.
</t1Density_threshold>
<t2Density_threshold> (float) [opt] upper integrated density
threshold
spot sizing
limit. Used in Seg2Dgel
segmenter.
</t2Density_threshold>
<t1Range_threshold> (float) [opt] lower density-range threshold
spot
sizing limit (maxDensity minDensity).
Used in
Seg2Dgel segmenter.
</t1Range_threshold>
<t2Range_threshold> (float) [opt] upper density-range threshold
spot
sizing limit (maxDensity minDensity). Used in
Seg2Dgel segmenter.
</t2Range_threshold>
<Percent_saturation_threshold> (float) [opt] minimum % spot
saturation
limit before invoke the spot slitting algorithm
during spot
segmentation. Used in Seg2Dgel segmenter.
</Percent_saturation_threshold>
<ccMinSize_threshold> (int) [opt] minimum central-core area size
of a
spot for it to be considered a spot, otherwise it
is assigned as
noise and removed. Used in Seg2Dgel segmenter.
</ccMinSize_threshold>
<Density_units> (String) [opt] grayscale calibration units if the
calibration
exists. Otherwise the default is
"grayscale"
</Density_units>
<Background_FilterSize> (int) [opt] size of the square zonal
notch-filter pixel
averaging
size used in estimating the background density map.
Used in Seg2Dgel segmenter.
</Background_FilterSize>
</Sample_parameters>
5.3.3 SSF spot data <Spot> schema
Each segmented spot is defined as a <Spot> object. There is
one spot instance for every segmented spot. Note that the following PEDRo field names are used here. Note that the optional fields are
defined in Seg2Dgel,
but are not required for other spot quantification programs.
<id>
<area>
<intensity>
<normalised_volume>
<volume>
<pixel_x_coord>
<pixel_y_coord>
<local_background>
The other fields were those required for Open2Dprot and that had
no PEDRo equivalent so we used our own names. This will be reconciled with the
new GelML/MIAPE standard.
<Spot>
<id> (int) the sequential spot identification number [1 : #spots
in
sample]. This is only unique within a
single SSF data set.
</id>
<area> (int) [opt] spot area A and is number of pixels in detected
spot </area>
<intensity> (float) integrated spot density, D, in the calibration
units
<Density_units> if the sample is calibrated (e.g., OD or
optical density, etc.), else sum of gray values. This
correlates with the amount of protein in the spot.
</intensity>
<normalised_volume> (float) density corrected for background or D
computed as (D = D A*mnDensity) in
<Density_units> if sample
is calibrated. If <local_background> is 0, then
has
same value as <intensity>.
</normalised_volume>
<volume>
(float) spot volume (in <Density_units> if the sample is
calibrated) computed as a gaussian volume:
sqrt(3.14159)*maxDensity*sxTot*syTot
If <maxDensity> is 0, then has same value as
the <normalised_volume>.
</volume>
<pixel_x_coord> (float) X centroid of segmented spot
</pixel_x_coord>
<pixel_y_coord> (float) Y centroid of segmented spot
</pixel_y_coord>
<local_background> (float) [opt] estimate of total background
density of
spot(estimated as A*mnDensity). 0 if not defined.
</local_background>
<minDensity> (float) [opt] minimum density within the spot (not
corrected
for background) in <Density_units> if the sample
is calibrated.
</minDensity>
<maxDensity> (float) [opt] maximum density within the spot (not
corrected for
background) in <Density_units> if the sample is
calibrated.
</maxDensity>
<meanDensity> (float) mean density/pixel within the spot (not
corrected for background) in <Density_units> if
sample is
calibrated. If <local_background> is 0, then has
same value
as <normalised_volume>.
</meanDensity>
<meanBackground> (float) [opt] mean background density/pixel of
the spot
or mnDensity in
<Density_units> if the sample is calibrated
</meanBackground>
<merX1> (int) [opt] left edge of spot minimum enclosing rectangle
</merX1>
<merY1> (int) [opt] top edge of spot minimum enclosing rectangle
</merY1>
<merX2> (int) [opt] right edge of spot minimum enclosing rectangle
</merX2>
<merY2> (int) [opt] bottom edge of spot minimum enclosing
rectangle</merY2>
<sxTot> (float) [opt] density-weighted spot size X standard
deviation in pixels
</sxTot>
<syTot> (float) [opt] density-weighted spot size Y standard
deviation in pixels
</syTot>
<sxyTot> (float) [opt] density weighted spot size covariance in
pixels
</sxyTot>
<ccNumber> (int) [opt] internal central core numbering used
by Seg2Dgel segmenter
</ccNumber>
<spotBoundaryStr> (String) [opt] chain-coded spot boundary if it
exists.
Codes: E=0, NE=1, N=2, NW=3, W=4, SW=5, S=6, SE=7.
</spotBoundaryStr>
</Spot>
5.3.4 SSF Epilogue statistics <Global_segmenter_statistics>
schema
These optional epilogue statistics are defined in the
<Global_segmenter_statistics> object. This includes information on the
total number, area, and density of spots accepted, rejected, etc. These
statistics are optional and are currently only used with the Seg2Dgel program.
<Global_segmenter_statistics>
<total_D_spots_accepted> the total number of spots D initially
accepted
before retesting after background correction
where (D = D A*mnDensity) for all spots.
</total_D_spots_accepted>
<total_densityPrime_spots_accepted> is statistics on D spots
accepted after threshold sizing.
</total_densityPrime_spots_accepted>
<total_omitted_densityPrime_spots_accepted is statistics on D
spots omitted after threshold sizing.
</total_omitted_densityPrime_spots_accepted>
<Pct_omitToAccept_densityPrime_spots_failing_t1Density_resizing>
(float) the ratio of omitted/accepted D spots failing
lower T1 density sizing threshold test
</Pct_omitToAccept_densityPrime_spots_failing_t1Density_resizing>
<Nbr_Spots_Failing_Area_Sizing> (float) numbers of spots failing
lower and upper area, density, and density range
threshold
sizing tests.
</Nbr_Spots_Failing_Area_Sizing>
</Global_segmenter_statistics>
Then, these complex types used in the above type are defined as
follows:
<total_D_spots_accepted>
<nbr> (int) number of D spots accepted</nbr>
<density>(float) total density of D spots accepted
</density>
<area> (int) total area of D spots
accepted </area>
</total_D_spots_accepted>
Then,
<total_densityPrime_spots_accepted>
<nbr> (int) number of D spots accepted </nbr>
<densityPrime>(float) total density of D spots accepted
</densityPrime>
<area> (int) total area of D spots accepted </area>
</total_densityPrime_spots_accepted>
Then,
<total_omitted_densityPrime_spots_accepted>
<nbr> (int) number of omitted D spots accepted </nbr>
<densityPrime>(float) total density of omitted D spots
accepted
</densityPrime>
<area> (int) total area of omitted D spots accepted </area>
</total_omitted_densityPrime_spots_accepted>
Then,
<Nbr_Spots_Failing_Area_Sizing>
<nbr_below_t1Area_thr>3316</nbr_below_t1Area_thr>
<nbr_above_t2Area_thr>0</nbr_above_t2Area_thr>
<nbr_below_t1Density_thr>4214</nbr_below_t1Density_thr>
<nbr_above_t2Density_thr>0</nbr_above_t2Density_thr>
<nbr_below_t1Range_thr>3923</nbr_below_t1Range_thr>
<nbr_above_t2Range_thr>0</nbr_above_t2Range_thr>
</Nbr_Spots_Failing_Area_Sizing>
5.4 Paired sample spot-list data
(SPF)
The Paired sample spot data (SPF) is created by pairing SSF
spot-lists from two different samples. For N samples (S1, S2,
, Sn), if you compare the same sample Sr with each of
the remaining N-1 samples, sample Sr is called the "reference
sample" or Rsample.
This allows you to use the transitivity relationship that if a
spot is known to be paired between Sr and Si, and the
spot in Sr is also paired with a spot in Sj, then that
spot (by transitivity) is paired between Si and Sj. Of
course, this does not take into account the possibility of false positive and
false negative pairing errors. That is a separate issue although it affects
the quality of the data.
The Composite Samples Database (CSD) uses this reference sample
design.
by requiring the declaration of a single reference sample used to
define a virtual-spot space in the database. Spots present in the Rsample are
assigned to Rspots sets of corresponding spots between samples. Spots missing
from the Rsample are extrapolated into the Rsample virtual-spot space and are
called extrapolated-Rspots (eRspot). SPF data using different reference samples
can still be merged into the CSD by identifying remapped Rsample Rspots or
ERspots. See Figure 1.a and the legend for more discussion of Rspot and ERspot.
Note: depending on the program that builds the Composite Samples
Database (CSD) described in Section 5.5, you may or may not require the Rsample
to be the same sample. As long as there is a transitive connection between all
samples, one could compute corresponding spot pairings using different reference
samples. However, this may increase the spot pairing errors. There are other
ways of doing this (pair all samples with all samples) that minimizes this
problem.
Paired sample data
A paired sample data set from two samples, (i.e., spots between samples
are paired). The Sample Paired-spot-list File (SPF) is created by pairing the
SSF data for an Rsample and Sample. It may also include the landmark set for
these samples extracted from the landmark DB. The samples are linked from the
accession database. We have one example of the module that produces this data, CmpSpots, which pairs two
SSF files given landmark data. The paired spot-list DTD contains lists of
paired spots and also parameters used in computing the spot list and statistics
on the spot features.
5.4.1 Fields in the sample paired-spot-list file XSD
(Open2Dprot-SPF.xsd)
This is defined using the classes O2Plib.db.DbPairedSamples.java,
O2Plib.db.DbSpot.java and O2Plib.db.DbSample.java.
Each sample will have a processed standardized spot list we call a
Sample Paired-spot-list File (SPF). The SPF DTD is available at
http://open2dprot.sourceforge.net/O2Plib/Open2Dprot-SPF.dtd
The SPF database XSD is available from the Open2Dprot Web site CVS
server at
http://cvs.sourceforge.net/viewcvs.py/open2dprot/schemas/Open2Dprot-SPF.xsd?rev=1.14&view=log
The Sample Paired-spot-List File (SPF) is saved as
<project-directory>/xml/<Sample>-SPF.xml
A SPF is described below.
Here is an example of an SPF XML file in the demonstration
directory generated by the CmpSpots program,
http://open2dprot.sourceforge.net/demo/xml/gel-HM-071-SPF.xml
(The Reference sample is gel-HM-019-SSF, and Sample is
gel-HM-071-SPF).
The CmpSpots program that generated this XML file is
described at
http://open2dprot.sourceforge.net/CmpSpots
The SPF is divided into three parts:
a) A preface containing sample
experiment information and spot-pairing
parameters
b) multiple instances of <Pspot>
data
c) an epilogue containing sample
spot-pairing statistics
5.4.2 SPF Preface <Pairing_parameters> schema
The sample experiment information and pairing parameters are given
in the <Pairing_parameters> object.
<Pairing_parameters>
<date> (String) the date the file was created </date>
<Open2Dprot_SPF_Version> (String) version of SPF schema
</Open2Dprot_SPF_Version>
<Project_directory> (String) path of project directory relative
to Open2Dprot installation directory or full path
if it is elsewhere
</Project_directory>
<Sample_Pairs_File> (String) base name of SPF output file
</Sample_Pairs_File>
<thrSP_threshold> (float) [opt] maximum distance (dP feature) to
use for a spot to be called a sure-
</thrSP_threshold>
<thrPP_threshold> (float) [opt] maximum distance (dP feature) to
use
for a spot
to be called a possible-pair
</thrPP_threshold>
<nbrAltLandmarks> (float) [opt] number of adjacent alternate
landmark
sets to check when doing secondary pairing
</nbrAltLandmarks>
<nbrLandmarks> (int)[opt] number of landmarks used for these two
samples
</nbrLandmarks>
<Rsample> is the Rsample experiment and parameters
</Rsample>
<Sample> is the sample experiment and parameters </Sample>
</Pairing_parameters>
5.4.2.1 Contents of the <Rsample> and <Sample> objects
in the SPF schema
These objects (used for both <Rsample> and <Sample>)
contain the same fields so will be reported only once here. They reflect the
two different samples being paired. The example is given below.
<Sample>
<Sample_Type> (String) either "Rsample" or
"Sample" </Sample_Type>
<Sample_Name> (String) same <Sample_Name> as for SSF file
</Sample_Name>
<Simple_FileName> (String) is SSF input file (could be image) file
name
without the path
</Simple_FileName>
<Simple_Pix_FileName> (String) SSF input file (image or virtual
image)
file name with path derived from the project
directory.
</Simple_Pix_FileName>
<Simple_SSF_FileName> (String) SSF output file input to
spot-pairing
program. The full file name path is derived from the
project directory.
</Simple_SSF_FileName>
<cwx1> (int) left edge of computing window in pixels </cwx1>
<cwx2> (int) right edge of computing window in pixels
</cwx2>
<cwy1> (int) top edge of computing window in pixels </cwy1>
<cwy2> (int) bottom edge of computing window in pixels
</cwy2>
<Pix_Height> (int) height of image in pixels for sample
</Pix_Height>
<Pix_Width> (int) width of image in pixels of sample
</Pix_Width>
<PixelSizeMicrons> (float) pixel size in microns for sample
</PixelSizeMicrons>
<NbrSpots> (float) number of spots available in SSF data set for
this sample
</NbrSpots>
<NbrSpotsPrime>
(int) number of valid D spots found in SSF data.
D indicates spot density D was corrected for
background
</NbrSpotsPrime>
<NbrSpotsOmitted> (int) number of invalid D spots found in SSF
data
</NbrSpotsOmitted>
<TotSampleDensity> (float) total density of all valid D spots
found in SSF data
</TotSampleDensity>
<TotSampleDensityPrime> (float) total density of all valid D
spots found
in the SSF data
</TotSampleDensityPrime>
<TotOmittedDensity> (float) total density of D spots omitted from
SSF data
</TotOmittedDensity>
<TotSampleArea> (float) total area of all valid D spots found in
the SSF data
</TotSampleArea>
<TotSampleAreaPrime> (float) total area of all valid D spots
found in SSF data
</TotSampleAreaPrime>
<TotSampleAreaOmitted> (float) total area of D spots omitted from
the SSF data
</TotSampleAreaOmitted>
</Sample>
The <RSample>
is defined the same as the <Sample> but the data is for the Rsample so it
is not shown here.
We now show examples for the <Rsample> and <Sample>
objects. Note that this SPF output data was inherited from the SSF input files.
The following is an example,
<Pairing_parameters>
<Rsample>
<Sample_Type>"Rsample"</Sample_Type>
<Sample_Name>"gel-HM-019"</Sample_Name>
<Simple_FileName>"gel-HM-019.gif"</Simple_FileName>
<Sample_Pix_FileName>"demo/ppx/gel-HM-019.gif"</Sample_Pix_FileName>
<Sample_SSF_FileName>"demo/xml/gel-HM-019-SSF.xml"</Sample_SSF_FileName>
<cwx1>14</cwx1>
<cwx2>475</cwx2>
<cwy1>74</cwy1>
<cwy2>509</cwy2>
<Pix_Height>512</Pix_Height>
<Pix_Width>512</Pix_Width>
<PixelSizeMicrons>0.00</PixelSizeMicrons>
<NbrSpots>1588</NbrSpots>
<NbrSpotsPrime>738</NbrSpotsPrime>
<NbrSpotsOmitted>850</NbrSpotsOmitted>
<TotSampleDensity>8838.70</TotSampleDensity>
<TotSampleDensityPrime>5670.40</TotSampleDensityPrime>
<TotOmittedDensity>91.90</TotOmittedDensity>
<TotSampleArea>36158</TotSampleArea>
<TotSampleAreaPrime>23019</TotSampleAreaPrime>
<TotSampleAreaOmitted>13139</TotSampleAreaOmitted>
</Rsample>
<Sample>
<Sample_Type>"Sample"</Sample_Type>
<Sample_Name>"gel-HM-071"</Sample_Name>
<Simple_FileName>"gel-HM-071.gif"</Simple_FileName>
<Sample_Pix_FileName>"demo\ppx\gel-HM-071.gif"</Sample_Pix_FileName>
<Sample_SSF_FileName>"demo\xml\gel-HM-071-SSF.xml"</Sample_SSF_FileName>
<cwx1>6</cwx1>
<cwx2>450</cwx2>
<cwy1>68</cwy1>
<cwy2>503</cwy2>
<Pix_Height>512</Pix_Height>
<Pix_Width>512</Pix_Width>
<PixelSizeMicrons>0.00</PixelSizeMicrons>
<NbrSpots>4530</NbrSpots>
<NbrSpotsPrime>2439</NbrSpotsPrime>
<NbrSpotsOmitted>2091</NbrSpotsOmitted>
<TotSampleDensity>54230.70</TotSampleDensity>
<TotSampleDensityPrime>40021.30</TotSampleDensityPrime>
<TotOmittedDensity>182.60</TotOmittedDensity>
<TotSampleArea>124076</TotSampleArea>
<TotSampleAreaPrime>84136</TotSampleAreaPrime>
<TotSampleAreaOmitted>37070</TotSampleAreaOmitted>
</Sample>
</Pairing_parameters>
5.4.3 SPF paired-spot <Pspot> schema
Most of the SPF file consists of paired spot instances using the
<Pspot> object. There is a <Pspot> for each spot pair and it is
assigned a pairing code <PairingCode> (SP, PP, and AP described in CmpSpots documentation).
There is also an entry for each unpaired spot in either sample (assigned a
pairing label US). Note that each paired or unpaired spot <Pspot> belongs
to a landmark set. For data where pairing codes are not relevant such as
already paired protein array data, these codes and landmark data could be
ignored.
<Pspot>
<LandmarkSet> (String) landmark set to which this spot pair
belongs. This can be
a number or letter.
</LandmarkSet>
<R_spotNbr> (int) spot number in Rsample spot list from SSF data
</R_spotNbr>
<R_dxLM> (int) X (Cartesian) offset of Rsample spot from landmark
</R_dxLM>
<R_dyLM> (int) Y (Cartesian) offset of the Rsample spot from
landmark </R_dyLM>
<R_xLM> (int) X centroid of Rsample spot from SSF data
</R_xC>
<R_yLM> (int) Y centroid of Rsample spot from SSF data
</R_yC>
<R_merX1> (int) left edge of the Rsample spot from the SSF data
</R_merX1>
<R_merX2> (int) right edge of the Rsample spot from SSF data
</R_merX2>
<R_merY1> (int) top edge of the Rsample spot from the SSF data
</R_merY1>
<R_merY21> (int) bottom edge of Rsample spot from SSF data
</R_merY2>
<S_spotNbr> (int) spot number in Sample spot list from SSF data </S_spotNbr>
<S_dxLM> (int) X (Cartesian) offset of Sample spot from landmark
</S_dxLM>
<S_dyLM> (int) Y (Cartesian) offset of the Sample spot from
landmark </S_dyLM>
<S_xLM> (int) X centroid of Sample spot from SSF data
</S_xC>
<S_yLM> (int) Y centroid of Sample spot from SSF data
</S_yC>
<S_merX1> (int) left edge of Sample spot from SSF data
</S_merX1>
<S_merX2> (int) right edge of Sample spot from SSF data
</S_merX2>
<S_merY1> (int) top edge of the Sample spot from the SSF data
</S_merY1>
<S_merY2> (int) bottom edge of the Sample spot from the SSF data
</S_merY2>
<PairingCode> (String) [opt] a letter pairing code: S
(SP=sure-pair),
P (PP= possible-pair), A (AP= ambiguous-pair), U (US=
unresolved-spot),
C (CP= CSD composite replicates pair), E (EP=
extrapolated-pair).
Used if spot-pairing program generates these types
of pairing codes
(see CmpSpots).
</PairingCode>
<DP> (float) [opt] the distance between the paired spots after
pairing.
Used if spot-pairing program generates this type of
data
(see CmpSpots).
</DP>
<DL> is the distance from the center of the paired spots to the
nearest landmark
set. Optional. Used if spot-pairing program generates
this type of
data (see CmpSpots).
</DL>
<R_area> (int) is the area of the Rsample spot</R_area>
<S_area> (int) is the area of the Sample spot </S_area>
<R_dens> (float) total density D of the Rsample spot
</R_dens>
<S_dens> (float) total density D of the Sample spot
</S_dens>
<R_dPrime> (float) total density D (background corrected) of
Rsample spot.
</R_dPrime>
<S_dPrime> (float) total density D (background corrected) of Sample spot.
</S_dPrime>
<R_volume> (float) volume of the Rsample spot</R_volume>
<S_volume> (float) volume of the Sample spot </S_volume>
<R_MaxDens> (float) maximum density/pixel of the Rsample spot
</R_MaxDens>
<S_MaxDens> (float) maximum density/pixel of the Sample spot
</S_MaxDens>
<R_MinDens> (float) minimum density/pixel of the Rsample spot
</R_MinDens>
<S_MinDens> (float) minimum density/pixel of the Sample spot
</S_MinDens>
<R_MeanBkgDens> (float) mean background density/pixel of Rsample
spot
</R_MeanBkgDens>
<S_MeanBkgDens> (float) mean background density/pixel of the
Sample spot
</S_MeanBkgDens>
<R_stdDev_X>
(float) density weighted X std deviation of Rsample spot
</R_stdDev_X>
<R_stdDev_Y> (float) density weighted Y std deviation of Rsample
spot
</R_stdDev_Y>
<S_stdDev_X> (float) density weighted X std deviation of Sample
spot
</S_stdDev_X>
<S_stdDev_Y> (float) density weighted Y std deviation of Sample
spot
</S_stdDev_Y>
<R_stdDev_Mean_X> (float) [opt] estimated mean X position in
Rsample Cspot
</R_stdDev_Mean_X>
<R_stdDev_Mean_Y> (float) [opt] estimated mean Y position in
Rsample Cspot
</R_stdDev_Mean_Y>
<R_stdDev_Density> (float) [opt] std deviation of Rsample Cspot
density.
</R_stdDev_Density>
<S_stdDev_Density> (float) [opt] std deviation of Sample Cspot
density.
</S_stdDevDensity>
<NbrSamples_in_CsamplePrime> (int) [opt] number of samples in the
Cspot.
Used if using CP pairing codes from Csample from CSD.
</NbrSamples_in_CsamplePrime>
</Pspot>
If a <Pspot> is part of a
<Csample> (see Section 5.5.1), then the data
fields contain the mean data and the additional fields <R_stdDev_Mean_X>,
<R_stdDev_Mean_Y>, <R_stdDev_Density>, <S_stdDev_Density, and
<NbrSamples_in_CsamplePrime> now contain data.
5.4.4 SPF Epilogue <Global_Spot_pairing_statistics> schema
The optional pairing statistics is defined in the
<Global_Spot_pairing_statistics> object instance. It consists of several
sections having to do with numbers of SP, PP, AP, US, etc. pairing labels after
primary pairing and then after Secondary pairing (see CmpSpots documentation).
These statistics are currently only associated with the CmpSpots program.
<Global_Spot_pairing_statistics>
<NbrRsampleSpotsInLMS> (int) [opt] number of Rsample spots
in landmark set.
Used if spot-pairing program generates this type of data (see CmpSpots).
</NbrRsampleSpotsInLMS>
<NbrSampleSpotsInLMS> (int) [opt] number of Sample spots in
the landmark set.
Used if spot-pairing program generates this type of data (see CmpSpots).
</NbrSampleSpotsInLMS>
<Landmark_set_sizes> (int) [opt]
list of landmark set sizes for the landmarks
in the landmark set. Used if spot-pairing program generates
this type of data (see CmpSpots).
</Landmark_set_sizes>
<InitialpairingStats> [opt] complex type holding pairing
statistics.
Used if spot-pairing program generates this type of data (see CmpSpots).
</InitialpairingStats>
<SecondarypairingStats> [opt] complex type holding pairing
statistics
after secondary pairing. Used if spot-pairing program generates this
type of data (see CmpSpots).
</SecondarypairingStats>
<Primary_SP_PP_pairRate> (float) the percent of all SP and PP
paired spots
after primary pairing.
</Primary_SP_PP_pairRate>
<Secondary_SP_PP_pairRate> (float) the percent of all SP and PP
paired spots
after secondary pairing
</Secondary_SP_PP_pairRate>
<meanDP_SP_PP> (float) mean DP for all SP and PP paired spots for
both samples
</meanDP_SP_PP>
<meanDPprime_SP_PP> (float) the adjusted mean DP for all SP and PP
paired
spots for the Rsample only
</meanDPprime_SP_PP>
</Global_Spot_pairing_statistics>
Then, the
<Landmark_set_sizes> list of landmark set sizes type is defined
as:
<Landmark_set_sizes>
<Landmark> landmark set 1 spot countsdata </Landmark>
<Landmark> landmark set 2 spot countsdata </Landmark>
. . .
<Landmark> landmark set n spot countsdata </Landmark>
</Landmark_set_sizes>
Then, the <Landmark> type is defined
as:
<Landmark>
<Landmark_name> (String) name of the landmark
</Landmark_name>
<Landmark_nbr> (int) landmark number index value
</Landmark_nbr>
<Nbr_Rsample_spots> (int) the number of Rsample spots in this
landmark set
</Nbr_Rsample_spots>
<Nbr_Sample_spots> (int) the number of Sample spots in this
landmark set
</Nbr_Sample_spots>
</Landmark>
Then, the <InitialpairingStats> is
defined as:
<InitialpairingStats>
<Nbr_US_spotsPri> (int) number of US
pairs found during primary pairing
</Nbr_US_spotsPri>
<Nbr_SP_spotsPri> (int) number of SP pairs found during primary
pairing
</Nbr_SP_spotsPri>
<Nbr_PP_spotsPri> (int) number of PP pairs found during primary
pairing
</Nbr_PP_spotsPri>
<Nbr_AP_spotsPri> (int) number of AP pairs found during primary
pairing
</Nbr_AP_spotsPri>
<Nbr_CP_spotsPri> (int) number of CP pairs found during primary
pairing
</Nbr_CP_spotsPri>
<Nbr_EP_spotsPri> (int) number of EP pairs found during primary
pairing
</Nbr_EP_spotsPri>
</InitialpairingStats>
Then, the <SecondarypairingStats>
complex type is defined as:
<SecondarypairingStats>
<Nbr_US_spotsSec> (int) number of US spots found after secondary
pairing
</Nbr_US_spotsSec>
<Nbr_SP_spotsSec> (int) number of SP pairs found after secondary
pairing
</Nbr_SP_spotsSec>
<Nbr_PP_spotsSec> (int) number of PP pairs found after secondary
pairing
</Nbr_PP_spotsSec>
<Nbr_AP_spotsSec> (int) number of AP pairs found after secondary
pairing
</Nbr_AP_spotsSec>
<Nbr_CP_spotsSec> (int) number of CP pairs found after secondary
pairing
</Nbr_CP_spotsSec>
<Nbr_EP_spotsSec> (int) number of EP pairs found after secondary
pairing
</Nbr_EP_spotsSec>
</SecondarypairingStats>
5.5 Composite Sample Database (CSD)
XSD schema (Open2Dprot-CSD.xsd)
The CSD consists of experiment information and data merged from
all of the samples. It contains sample experiment information as well as spot
expression data of a set of paired spots (that can be constructed from paired
spot data). See Figure 1.a and the legend for more discussion on the CSD. Note
that by paired spots in the context of the CSD, we mean spots paired to
the CSD virtual reference sample.
NOTE: we are in the process
of defining the XML schema for the CSD and so dont have the exact XML schema
we will use. However, we outline of some of the key export data of an assembled
CSD as define in this Section.
An early database XSD of part of the CSD schema is available from
the Open2Dprot Web site CVS server at
http://cvs.sourceforge.net/viewcvs.py/open2dprot/schemas/Open2Dprot-CSD.xsd?rev=1.14&view=log
The following CSD schema is being integrated into the Open2Dprot
CSD.
The <CSD> defines the CSD database. It is created
dynamically so that some components will not be present. It is defined in the
Java code from O2Plib.db.CSD.CSDio.writeXMLfile()
<CSD>
<CSD_name> name of the CSD DB </CSD_name>
<CreationDate> date CSD DB was created </CreationDate>
<EditDate> date CSD DB was changed or edited </EditDate>
<CSD_version> version of the CSD DB software </CSD_version>
<AccessionDB> name of the accession DB </AccessionDB>
<LandmarkDB> name of the landmark DB </LandmarkDB>
<Rsample> reference sample </Rsample>
<Totals> totals of
data in the CSD </Totals>
<Sizes> maximum
sizes allowed in the CSD </Sizes>
<MasterRspotList>
master Rspot list </MasterRspotList>
<Condition> set of
all samples in the CSD database </Condition>
<Condition> working
subset of samples in CSD [opt] </Condition>
<Condition> X
subset of samples in the CSD [opt] </Condition>
<Condition> Y
subset of samples in the CSD [opt] </Condition>
<Condition> Y
subset of samples in CSD [opt] </Condition>
<Condition>
expression profile list of samples [opt] </Condition>
<RspotList> user
defined Rspot list 1 [opt] </RspotList>
<RspotList> user
defined Rspot list 2 [opt] </RspotList>
. . .
<RspotList> user
defined Rspot list n [opt] </RspotList>
<RspotMap> user
defined Rspot map 1 [opt] </RspotMap>
<RspotMap> user
defined Rspot map 2 [opt] </RspotMap>
. . .
<RspotMap> user
defined Rspot map n [opt] </RspotMap>
<Condition> user
defined subset 1 of samples [opt] </Condition>
<Condition> user
defined subset 2 of samples [opt] </Condition>
. . .
<Condition> user
defined subset n of samples [opt] </Condition>
. . . (additional
objects) . . .
</CSD>
The following complex schema types help describe the data in above
<CSD> schema definition.
The <RspotList> defines the data for an list of <Rspot>s. It is defined in
the Java code from O2Plib.db.CSD.CSDRspotList.toXML(). As the result of
a CSD analysis, subsets of the Rspots in the total CSD can be computed. These
are useful for a variety of purposes including normalization, data filtering,
clustering, statistical tests, etc. E.g., an Rspot set may be the result of
some sort of clustering to group spots together with similar property or
properties (e.g., expression profile, function, etc.)
<RspotList>
<idRSL> (int) index of the Rspot List </idRSL>
<name> name of the Rspot list </name>
<timeStamp> timeStamp </timeStamp>
<title> title
</title>
<totRspots> (int) total number of Rspots </totRspots>
<nbrRspots> (int) number of Rspots </nbrRspots>
<nbrERspots> (int) number of ERspots </nbrERspots>
<Rspot> Rspot 1</Rspot>
<Rspot> Rspot 2</Rspot>
. . .
<Rspot> Rspot nbrRspots</Rspot>
<ERspot> ERspot 1</ERspot>
<ERspot> ERspot
2</ERspot>
. . .
<ERspot> ERspot nbrERspots</ERspot>
</RspotList>
The <Rspot> defines the data for an Rspot and consists of quantitative and
qualitative data (<Sample>, <Annotation>). It is defined in the
Java code from O2Plib.db.CSD.CSDRspot.toXML()
<Rspot>
<idNbr> idNbr (int) </idNbr>
<idRsampleSpot> (int) id of spot in Rsample </idRsampleSpot>
<xRspot> (float) centroid of Rspot </xRspot>
<yRspot> (float) centroid of Rspot </yRspot>
<eRspotFlag> (boolean) set if Rspot is an ERspot flag
</eRspotFlag>
<xERspot> (float) centroid of ERspot </xERspot>
<yERspot> (float) centroid of ERspot </yERspot>
<Annotation> annotation to use </Annotation>
<Normalization> normalization to use </Normalization>
<Sample> sample 1 </Sample>
<Sample> sample 2 </Sample>
. . .
<Sample> sample 1 </Sample>
<nSpots> (int) number of spots </nSpots>
<Pspot> paired spot 1 </Pspot>
<Pspot> paired spot 2 </Pspot>
. . .
<Pspot> paired spot n </Pspot>
</Rspot>
The <FullRspot> defines the full data for an Rspot and consists of quantitative
and qualitative data (<Sample>, <Annotation>). The
<FullRspot> is used by the <MasterRspotList>. It is defined in the
Java code from O2Plib.db.CSD.CSDRspot.toXML()
<FullRspotList>
<Rspot> full details on the Rspot </Rspot>
</FullRspotList>
The <Sample> defines the data for a Sample in the
CSD. This is a subset of the definition used in the SPF. It is defined in the
Java code from O2Plib.db.DbSample.toXML()
<Sample>
<nbr> (int) sample number index) </nbr>
<SampleName>
(String) sample name </SampleName>
</Sample>
The <Pspot> defines the quantitative spot features
for a spot-pair. The data is for Rsample spot and a sample spot. It is defined
in the Java code from O2Plib.db.DbPspot.toXML()
<Pspot>
*** NOTE <Pspot> is defined as part of the SPF in Section 5.4 ***
</Pspot>
The <Normalization> defines a normalization method
for a <RspotList> and <Rspot>. This is optional. It is defined in
the Java code from O2Plib.db.CSD.CSDnorm.toXML()
<Normalization>
<Name> (String) name of normalization </Name>
<Sample> (String) sample name </Sample>
<curNormMethod> (String) current normalization method
</curNormMethod>
( to be extended )
</Normalization>
The <Annotation> defines an annotation set for an
<Rspot>. This is optional. It is defined in the Java code from
O2Plib.db.CSD.CSDannotation.toXML()
<Annotation>
<Nbr> (int) annotation number (int) </Nbr>
<UniProtID> (String) Uniprot ID </UniProtID>
<SPID> (String) Swiss-Prot ID </SPID>
<SP-Name> (String) Swiss-Prot name </SP-Name>
( to be extended )
</Annotation>
The <Condition> defines a condition set of
<Sample>s. This is optional. It is defined in the Java code from
O2Plib.db.CSD.CSDcond.toXML(). Conditions are defined as a subset or
sub-list group of samples. Subsets of samples could be created as a result of
an analysis (such as a classifier) or created manually.
<Condition>
<idCond> (int) id number of condition </idCond>
<name> (String) name of condition </name>
<timeStamp> time stamp </timeStamp>
<title> (String) title of condition </title>
<nSamples> (int) number of samples n </nSamples>
<Sample> sample 1 </Sample>
<Sample> sample 2 </Sample>
. . .
<Sample> sample n </Sample>
</Condition>
The <Calibration> defines the grayscale and area
calibrations. These are optional. It is defined in the Java code from O2Plib.db.CSD.CSDcal.toXML()
<Calibration>
<calibName> (String) calibration name </calibName>
<calibType> (String) calibration
type </calibType>
<curveFittingType> (String) curveFittingType
</curveFittingType>
<areaCalibFlag> (boolean) use area Calibration
</areaCalibFlag>
<useCalibFlag> (boolean) use Calibration </useCalibFlag>
<pixelResolutionMicrons> pixel resolution microns
</pixelResolutionMicrons>
<pixXsize> (int) pix X size in pixels </pixXsize>
<pixYsize> (int) pix Y size in pixels </pixYsize>
<xName> (String) name of X axis </xName>
<xUnits> (String) units of X axis </xUnits>
<yName> (String) name of Y axis </yUnits>
<nXRspots> (int) nXRspots </nXRspots>
<nYRspots> (int) nYRspots </nYRspots>
<maxCalibrations> (int) maxCalibrations n
</maxCalibrations>
<CalibPoint> calibration point 1 </CalibPoint>
<CalibPoint> calibration point 2 </CalibPoint>
. . .
<CalibPoint> calibration point n </CalibPoint>
<exposureCorrectionFactor> (float) exposure correction factor
</exposureCorrectionFactor>
( to be extended )
</Calibration>
The <CalibPoint> defines a point in the
<Calibration>. It is defined in the Java code from
O2Plib.db.CSD.CSDcal.toXML()
<CalibPoint>
<nbr> calibration point number </nbr>
<xCoord> (int) x pixel coordinate </xCoord>
<yCoord> (int) y pixel coordinate </yCoord>
<rspot_Xcalib> (float) x calibrated coordinate
</rspot_Xcalib>
<rspot_Ycalib> (float) y calibrated coordinate
</rspot_Ycalib>
</CalibPoint>
The <LandmarkDB> defines the landmark database. It is
defined in the Java code from O2Plib.db.CSD.CSDlms.toXML()
<LandmarkDB>
<landmarkFile> (String) landmark DB file </landmarkFile>
<nLMsetsList> (int) # landmark sets n </nLMsetsList>
<LMsetData> landmark set 1 data
</LMsetData>
<LMsetData> landmark set 2 data
</LMsetData>
. . .
<LMsetData> landmark set n data
</LMsetData>
</LandmarkDB>
The <LMsetData> defines a landmark set data composed
of id, name and <LMset>. It is defined in the Java code from
O2Plib.db.Lmset.toXML()
<LMsetData>
<nbr> (int) landmark set number </nbr>
<lmSetName> (String) landmark set name </lmSetName>
<LMset> landmark set </LMset>
</LMsetData>
The <LMset> defines a landmark set composed of
<LM>s. Optional fields are indicated with [opt]. It is defined in the
Java code from O2Plib.db.Lmset.toXML()
<LMset>
<Rsample> (String) reference sample (Rsample) name </RsampleName>
<Sample> (String) sample name </SampleName>
<nbrLandmarks> (int) number of landmarks n </nbrLandmarks>
<editRsampleLMSFlag> (boolean) edit Rsample LMS [opt]
</editRsampleLMSFlag>
<LM> landmark number 1 </LM>
<LM> landmark number 2 </LM>
. . .
<LM> landmark number n </LM>
</LMset>
The <LM> defines the landmark database. Optional
fields are indicated with [opt]. It is defined in the Java code from
O2Plib.db.Lmset.toXML()
<LM>
<nbr> (int) landmark ID i </nbr>
<rLMx> (int) Rsample X coordinate </rLMx>
<rLMy> (int) Rsample Y coordinate </rLMy>
<sLMx> (int) sample X coordinate </sLMx>
<sLMy> (int) sample Y coordinate </sLMy>
<useLM> (Boolean) valid landmark flag [opt] </useLM>
<minEffectiveRadius> (int) min effective LM radius
[opt]</minEffectiveRadius>
<RdistNearestLM> (int) distance nearest Rsample LM [opt]
</RdistNearestLM>
<SdistNearestLM> (int) distance nearest sample LM [opt]
</SdistNearestLM>
<RdistNextNearestLM> (int) distance next nearest Rsample LM [opt]
</RdistNextNearestLM>
<SdistNextNearestLM> (int)
distance next nearest sample LM [opt]
</SdistNextNearestLM>
<RlmErrDist> (int) landmark error distance Rsample LM
</RlmErrDist>
<SlmErrDist> (int) landmark error distance sample LM
</SlmErrDist>
<RnearestLM> (int) nearest Rsample LM index </RnearestLM>
<SnearestLM> (int) nearest sample LM index </SnearestLM>
<RnextNearestLM> (int) next nearest Rsample LM [opt]
</RnextNearestLM>
<SnextNearestLM> (int) next nearest sample LM [opt]
</SnextNearestLM>
<RsetSizeLM> (int) size of Rsample LM set for LM [opt]
</RsetSizeLM>
<SsetSizeLM> (int) size of sample LM set for LM [opt]
</SsetSizeLM>
</LM>
The <MasterRspotSet> defines the CSD master Rspot set
list which is the list of all Rspots. Optional fields are indicated with [opt].
It is defined in the Java code from O2Plib.db.CSD.CSDRspotList.toXML()
<MasterRspotSet>
<idRSL> (int) index of the Rspot List </idRSL>
<name> name of the Rspot list </name>
<timeStamp> timeStamp </timeStamp>
<title> title
</title>
<totRspots> (int) total number of Rspots </totRspots>
<nbrRspots> (int) number of Rspots </nbrRspots>
<nbrERspots> (int) number of ERspots </nbrERspots>
<FullRspot> Rspot 1</FullRspot>
<FullRspot> Rspot 2</FullRspot>
. . .
<FullRspot> Rspot nbrRspots</FullRspot>
<FullERspot> ERspot 1</FullERspot>
<FullERspot> ERspot 2</FullERspot>
. . .
<FullERspot> ERspot nbrERspots</ERspot>
</MasterRspotSet>
The <Sizes> defines the CSD database total
statistics. Optional fields are indicated with [opt]. It is defined in the Java
code from O2Plib.db.CSD.CSDsizes.toXML()
<Sizes>
<maxCalib>
maxCalib </maxCalib>
<maxConditions>
(int) max number conditions allowed </maxConditions>
<maxImageCols>
(int) max virtual or real image columns </maxImageCols>
<maxImageRows>
(int) max virtual or real image rows </maxImageRows>
<maxFailcodes>
(int) max Fail codes [opt]</maxFailcodes>
<maxSamples>
(int) max number of landmark sets </maxLms>
<maxRspots>
(int) max number of Rspots allowed </maxRspots>
<maxFspots>
(int) max number for foreign Fspots allowed [opt]
</maxFspots>
<maxRspotBitSetFeatures>
(int) max Rspot Bit-set features
</maxRspotBitSetFeatures>
<maxRSLs> (int)
max number of RspotLists </maxRSLs>
<maxNodesSaftyFactor> max cache nodes safety factor [opt]
</maxNodesSaftyFactor>
<minRspotCacheBlocks>
(int) min size of Rspot Cache Blocks [opt]
</minRspotCacheBlocks>
<wrdsPerBlock>
(int) words per cache node [opt] </wrdsPerNode>
<estBufsize>
(int) estimated buffer size [opt] </estBufsize>
<cacheBufSizeBlocks>
(int) cache buffer size blocks [opt]
</cacheBufSizeBlocks>
<maxNodesPerRspotSet> (int) max cache nodes per Rspot set [opt]
</maxNodesPerRspotSet>
<maxFileSize>
(int) max cache file size [opt] </maxFileSize>
</Sizes>
The <Totals>, which is optional, defines the CSD
database total statistics. Optional fields are indicated with
[db.CSD.CSDtotalsdb.Lmset.toXML()
<Totals>
<areaMax>
(int)
spot area Max [opt] </areaMax>
<areaMin>
(int)
spot area Min [opt] </areaMin>
<dLmax>
(int)
spot pairing dL max [opt] </dLmax>
<dPmax>
(int)
spot pairing dP max [opt] </dPmax>
<dPrimeMax> (int)
spot D
Max [opt] </dPrimeMax>
<dPrimeMin> (int)
spot D
Min [opt] </dPrimeMin>
<volumeMax> (int)
spot volume
Max [opt] </volumeMax>
<volumeMin> (int)
spot volume
Min [opt] </volumeMin>
<maxDmax> (int)
max
pixel density Dmax [opt] </maxDmax>
<minDmax> (int)
min
pixel density Dmax [opt] </minDmax>
<nbrLblSP> (int)number
of pairing labels of type SP [opt] </nbrLblSP>
<nbrLblPP> (int)number
of pairing labels of type PP [opt] </nbrLblPP>
<nbrLblAP> (int)number
of pairing labels of type AP [opt] </nbrLblAP>
<nbrLblUS> (int)number
of pairing labels of type US [opt] </nbrLblUS>
<nbrLblEP> (int)number of pairing
labels of type EP [opt] </nbrLblEP>
<nbrLblCP> (int)number of pairing
labels of type CP [opt] </nbrLblCP>
<nbrLblGP> (int)number of pairing labels
of type GP [opt] </nbrLblGP>
<totSampleDensity> (float) total sample density [opt] </totSampleDensity>
<totSampleArea> (float)
total
sample area [opt] </totSampleArea>
<totSampleNbrSpots> (int) total sample number of spots [opt]
</totSampleNbrSpots>
<totSampleDensityPrime> (float) total sample D spots [opt]
</totSampleDensityPrime>
<totSampleAreaPrime> (float) total sample area D spots [opt]
</totSampleAreaPrime>
<totSampleNbrSpotsPrime> (int) total sample number D spots [opt]
</totSampleNbrSpotsPrime>
<omittedDensity> (float) omitted spots
density [opt] </omittedDensity>
<omittedArea> (float) omitted spots area
[opt] </omittedArea>
<omittedNumberSpots> (int) omitted number spots [opt] </omittedNumberSpots>
<maxCalibGrayFound> (int) max calibrated gray value found [opt]
</maxCalibGrayFound>
<maxAreaFound> (float) max spot area
found [opt] </maxAreaFound>
<maxDensityFound> (float) max spot density found [opt] </maxDensityFound>
<maxDensityPrimeFound> (float) max spot D found [opt]
</maxDensityPrimeFound>
<invalidCentroidsTot> (int) number invalid centroids total [opt]
</invalidCentroidsTot>
<nSpotsAborted> (int) number spots
aborted [opt] </nSpotsAborted>
<ratioOmittedToAccepted> (float) ratio omitted D/accepted D [opt]
</ratioOmittedToAccepted>
<ctrArea1Failed> (int) ctrArea1Failed [opt]
</ctrArea1Failed>
<ctrArea2Failed> (int) ctrArea2Failed [opt]
</ctrArea2Failed>
<ctrDensity1Failed> (int) ctrDensity1Failed [opt]
</ctrDensity1Failed>
<ctrDensity2Failed> (int) ctrDensity2Failed [opt]
</ctrDensity2Failed>
<ctrDR1Failed> (int) ctrDR1Failed [opt] </ctrDR1Failed>
<ctrDR2Failed> (int) ctrDR2Failed [opt] </ctrDR2Failed>
<minBkgrdDensity> (float) min background density [opt] </minBkgrdDensity>
<maxBkgrdDensity> (float) max background
density [opt] </maxBkgrdDensity>
<minBkgrdTot> (float) min background
density total [opt] </minBkgrdTot>
<maxBkgrdTot> (float) max background
density total [opt] maxBkgrdTot>
</Totals>
5.5.1 The canonical sample,
Csample, for replicate samples in the CSD
Because databases could be built using huge numbers of samples and their sample spot lists, we offer the opportunity to collapse subsets of the CSD consisting of replicate samples into a statistical representation of those samples we call a canonical sample or Csample which is an estimate of the mean values for the samples. This is visualized in Figure 1.b. Each set of replicate samples could collapse the corresponding data into a separate canonical sample. For example, for k samples, we estimate Csample virtual spots coordinates in the virtual reference sample coordinate space as the mean spot positions of corresponding spots for those k samples. Similarly, it estimates other spot features such as area, density D, density D, min density, max density, etc. It also tracks for each spot the number of samples present for that spot so as to handle missing spot data.
The <Csample> is similar to Sample, but contains a list of the samples from which it was created. Essentially, the <Csample> is a <Condition> list of samples that was compressed into a single virtual sample.
<Csample>
<CsampleName> (String) name of the composite sample </CsampleName>
<NbrSamples> (int) number of samples n </NbrSamples>
<SampleName> name of sample 1</Sample>
<SampleName> name of sample 2</Sample>
. . .
<SampleName> name of sample n</Sample>
<Sample_parameters> Mean Csample info and parameters
</Sample_parameters>
<Global_segmenter_statistics> summary Csample statistics
</Global_segmenter_statistics>
</Csample>
The <Rspots> constructed from a set of <Samples>
5.5.2 The expression data for a single spot in the CSD or Rspot
A single CSD spot, which we call an Rspot, contains the expression
values across a set of samples for the spot paired across samples.
It could contain two types of data: unnormalized and an optional
normalized data. Note that unnormalized data is calibrated data as determined
in the SSF generation process. The reason you want both is that you may let the
investigator try various normalization methods some of which may require
unnormalized or uncalibrated data. So one must always be able to get back to
the primary data.
Spots which are missing in the reference sample (Rsample) but
present in the other samples may be extrapolated into the CSD virtual reference
sample space as extrapolated Rspots or ERspots (which could be assign an EP
pairing label). Various data-mining searchers could then be performed on these
types of virtual spots.
In addition, we can collapse replicate samples into composite
spots
or Cspots (with a CP pairs label) as discussed for
<Csample>s above. A composite spot is a statistical entity with numbers
of samples, mean and standard deviation of expression density and area and
other features used in place of the single sample data.
5.5.3 A post-translational modification Rspot or <PTMRspot>
To take post-translational modifications (PTM) into account, a
<PTMRspot> consisting of a set of Rspots could be defined. This allows
flexibility in treating each PTM as a separate spot or to sum the expression
for each of the variants. The CSD could handle this using the <RspotList>
complex type. We may want define a <PTMRspot> as an extended
<RspotList> with additional properties that could be used in additional
data-mining searches taking specific properties of the group of PTM spots into
account.
5.5.4 The <ConditionsList> as a list of <Conditions>
groups of samples
Just as there are sets or lists (ordered sets) of Rspots
<RspotList> and condition sets or lists of samples <Condition>,
there are ordered lists of conditions (e.g., lists of conditions sets of
replicate samples, a time series of replicates at each time point, or a
drug-dose response of replicates, etc). This type of structure lets us compare
mean expression profiles as defined by the higher order list of conditions. The
latter <ConditionsList> is defined as:
<ConditionsList>
<idCondList> (int) id number of condition list </idCondList>
<name> (String) name of condition list </name>
<timeStamp> time stamp </timeStamp>
<title> (String) title of condition list </title>
<nConditions> (int) number of samples n </ nConditions >
<Condition> condition 1 </Condition>
<Condition> condition 2 </Condition>
. . .
<Condition> condition n </Condition>
</ConditionsList>
--- end of document ---